Table of Contents
Definition / general | Essential features | Uses by pathologists | Clinical features | Case reports | Treatment | Microscopic (histologic) images | Immunohistochemistry & special stainsCite this page: Younes S. MSH2. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/stainsmsh2.html. Accessed March 31st, 2025.
Definition / general
- Protein is 104.7 kDa, 943 amino acids, located at chromosome 2p21
- Formed of 16 exons, mRNA is 3145 base pairs
Essential features
- One of the main genes of the MisMatch Repair family (MMR)
- The four key genes identified to date are MutL Homologue 1 (MLH1), MutS Homologue 2 (MSH2), MutS Homologue 6 (MSH6) and PostMeiotic Segregation increased 2 (PMS2, homologous to the E. coli MMR genes)
- Optimally functioning MMR is important for cancer avoidance and to maintain genomic stability by correcting single base mismatch and insertion deletion loops which results from DNA replication, genetic recombination or chemical or physical damage
- The MSH2 and MSH6 proteins bind, forming a heterodimeric complex (mutSα) which identifies mismatched bases and initiates DNA repair
- Mismatch binding results in an ATP dependent conformational change, with subsequent recruitment of mutLα, MLH1 and PMS2 heterodimers
- Then DNA repair proteins are recruited to complete the process
- MMR dysfunction causes microsatellite instability, which is characteristic feature of Lynch syndrome
- Microsatellites are short tandem repeats (2-5 DNA bases) scattered mainly in noncoding DNA regions
- Microsatellite instability (MSI) is defined as changes in length due to insertions or deletions of repeating units in a microsatellite; it is caused by defective MMR, and classified as MSI-H if more than 30-40% of investigated loci show MSI or MSI-L if less
Uses by pathologists
-
MSH2 and disease
- Lynch syndrome, hereditary non-polyposis colon cancer (HNPCC)
- Autosomal dominant
- Early age of onset
- Familial predisposition
- Colorectal carcinoma, gynecological and urological malignancies
- Molecular definition is based on germline mutation in one of the 4 major genes forming MMR system, 40% is caused by MSH2
- Diagnosis by
- Amsterdam criteria (Gastroenterology 1999;116:1453)
- Bethesda Guidelines / US National Cancer Institute (J Natl Cancer Inst 1997;89:1758)
- Revised Bethesda Guidelines (J Natl Cancer Inst 2004;96:261)
- Jerusalem criteria, recommending that either MMR IHC or MSI testing be carried out on every colorectal tumor if the patient is <70 years at diagnosis (Gastroenterology 2010;138:2197)
- Muir-Torre syndrome
- Autosomal dominant
- Sebaceous neoplasms and visceral malignancies
- Mainly caused by MSH2 mutation
- MMR defect and sporadic colorectal cancer
- 10 and 15% of sporadic colon cancers; this MSI phenotype is associated with proximal primary tumor location, high grade, mucinous pathology, early stage, females, smoking and older age at onset
- Most are thought to arise from sessile serrated adenomas or polyps
- The majority are caused by MLH1 methylation and inactivation
Clinical features
- MSH2 downregulation is associated with hepatocarcinogenesis (Cancer Res 2016 Jun 3 [Epub ahead of print])
- MSH2 RNA expression is deregulated in uterine fibroid and its protein expression level is increased in fibroid compared to normal myometrium (Reprod Sci 2016 Mar 31 [Epub ahead of print])
- MSH2 polymorphism is related to early onset of breast cancer in Taiwan (Ann Surg Oncol 2016 Mar 14 [Epub ahead of print])
Case reports
- 54 year old woman with familial adrenocortical carcinoma resulting from MSH2 mutation and Lynch syndrome (J Clin Endocrinol Metab 2016;101:2269)
Treatment
- Baicalein is a new molecule that is found to specifically shrinks MutSa-deficint xenografts tumor and inhibits growth of colon tumors in colon specific MSH2 knockout mice (Cancer Res 2016 Jun 4 [Epub ahead of print])
Microscopic (histologic) images
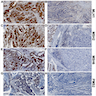
Immunohistochemistry & special stains
- Staining localization
- Nuclear
- MSH2 staining
- IHC is a simple way to detect protein expression
- Defective MMR will show loss of expression of the protein