Table of Contents
Definition / general | Essential features | Pathophysiology | Clinical features | Interpretation | Uses by pathologists | Prognostic factors | Microscopic (histologic) images | Positive staining - normal | Positive staining - disease | Negative staining | Molecular / cytogenetics description | Molecular / cytogenetics images | Sample pathology report | Board review style question #1 | Board review style answer #1 | Board review style question #2 | Board review style answer #2Cite this page: Alikhan M. TFE3. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/stainsTFE3.html. Accessed April 1st, 2025.
Definition / general
- TFE3 gene, located on Xp11.2, is translocated with > 20 of partner genes in a variety of tumor types, most commonly in MiT family translocation renal cell carcinoma (RCC)
- Translocation often leads to upregulation of TFE3 and subsequent cell proliferation and oncogenesis
- TFE3 gene translocation results in aberrant overexpression of the fusion oncoprotein which can be detected by a C terminus binding site TFE3 antibody (Adv Anat Pathol 2022;29:131)
Essential features
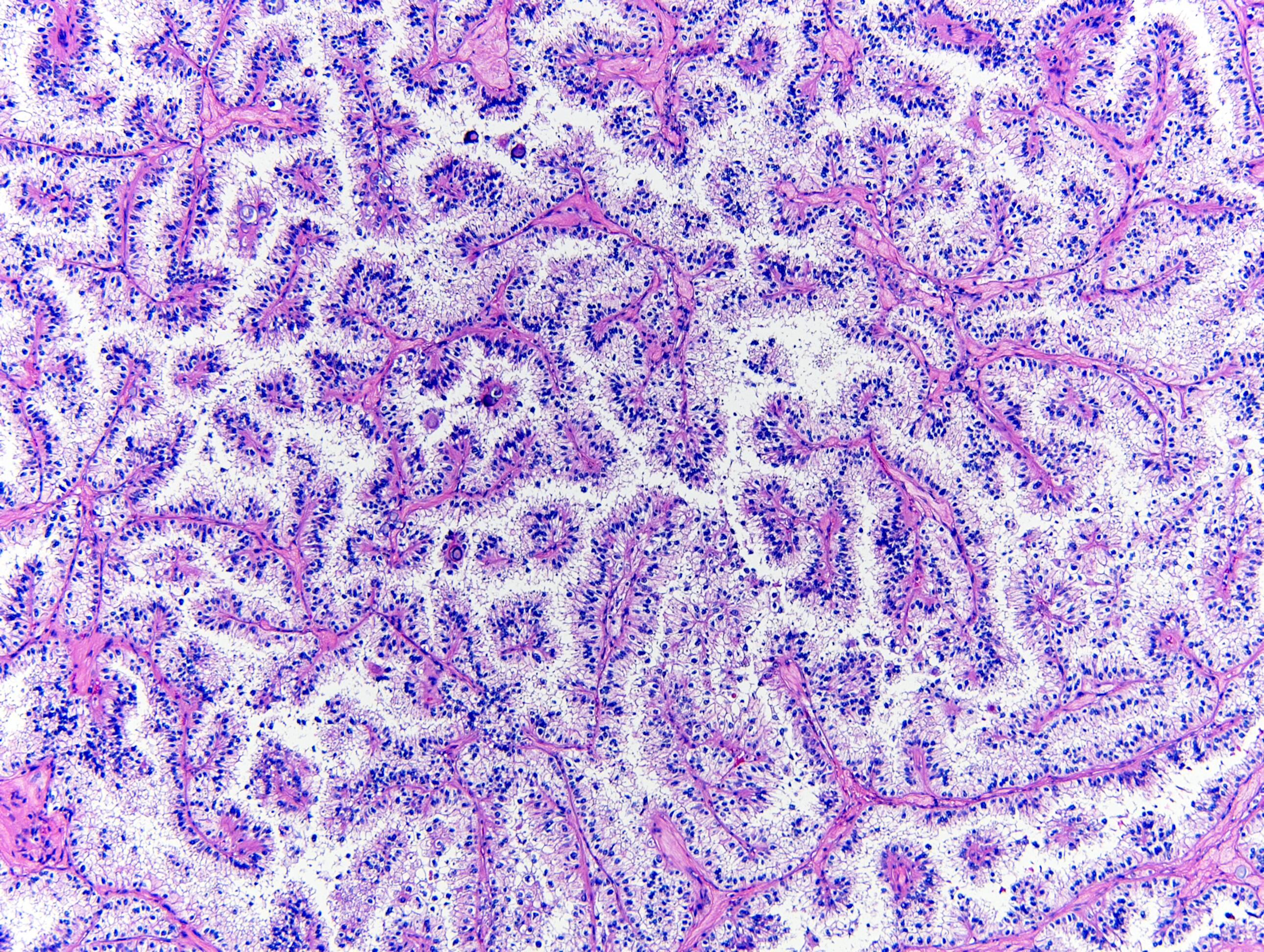
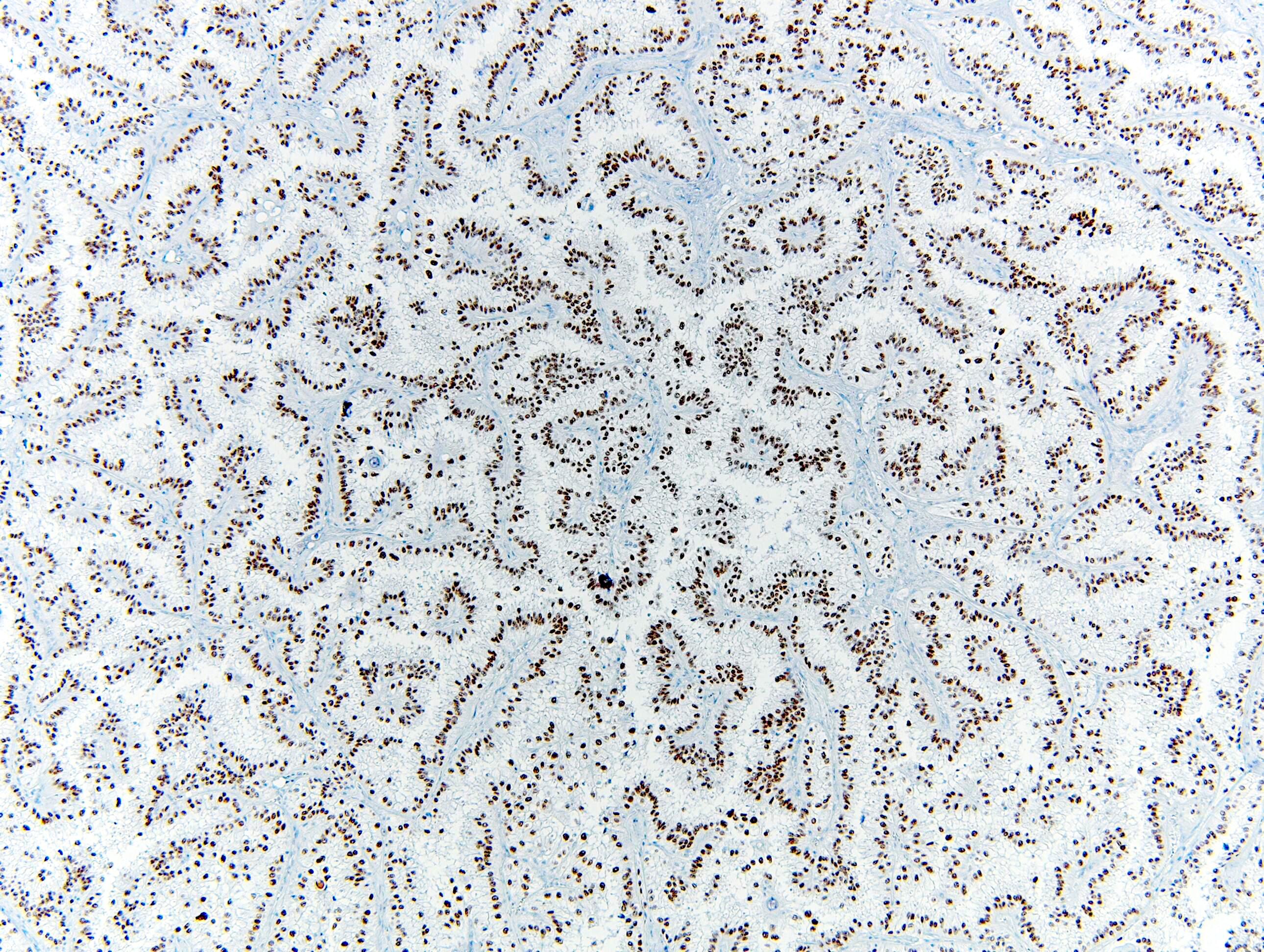
- Nuclear expression seen in MiT family translocation associated renal cell carcinoma (Am J Surg Pathol 2009;33:1894)
- Also observed in other neoplasms with translocations involving the TFE3 gene, such as alveolar soft part sarcoma t(X;17)(p11;q25) (Histopathology 1993;23:439)
- Subset of other tumors may also overexpress TFE3 by immunohistochemistry:
- Epithelioid hemangioendotheliomas with YAP1-TFE3 t(X;11)(p11;q22) (Genes Chromosomes Cancer 2013;52:775)
- Perivascular epitheloid cell tumors (PEComas) with TFE3 gene fusions (Am J Surg Pathol 2010;34:1395)
- Subset of solid pseudopapillary neoplasms of the pancreas and ovarian sclerosing stromal tumor (Am J Clin Pathol 2017;149:67, Anticancer Res 2017;37:5441)
- These are thought to not carry TFE3 associated translocations and likely harbor an alternate mechanism of TFE3 overexpression
Pathophysiology
- TFE3 gene encodes a transcription factor
- As a homodimer or heterodimer with MiT related proteins (such as TFEB), it promotes the expression of a wide variety of genes involved in cell growth and proliferation
- These proteins also bind DNA transcription enhancing sites to regulate cellular catabolism and nutrient dependent lysosomal response (Nat Commun 2019;10:3623)
- Due to their association with MiT, they may express melanocytic markers and downregulate expression of cytokeratins
Clinical features
- MiT family translocation renal cell carcinoma, like many other MiT related tumors, is more commonly seen in children and young adults
Interpretation
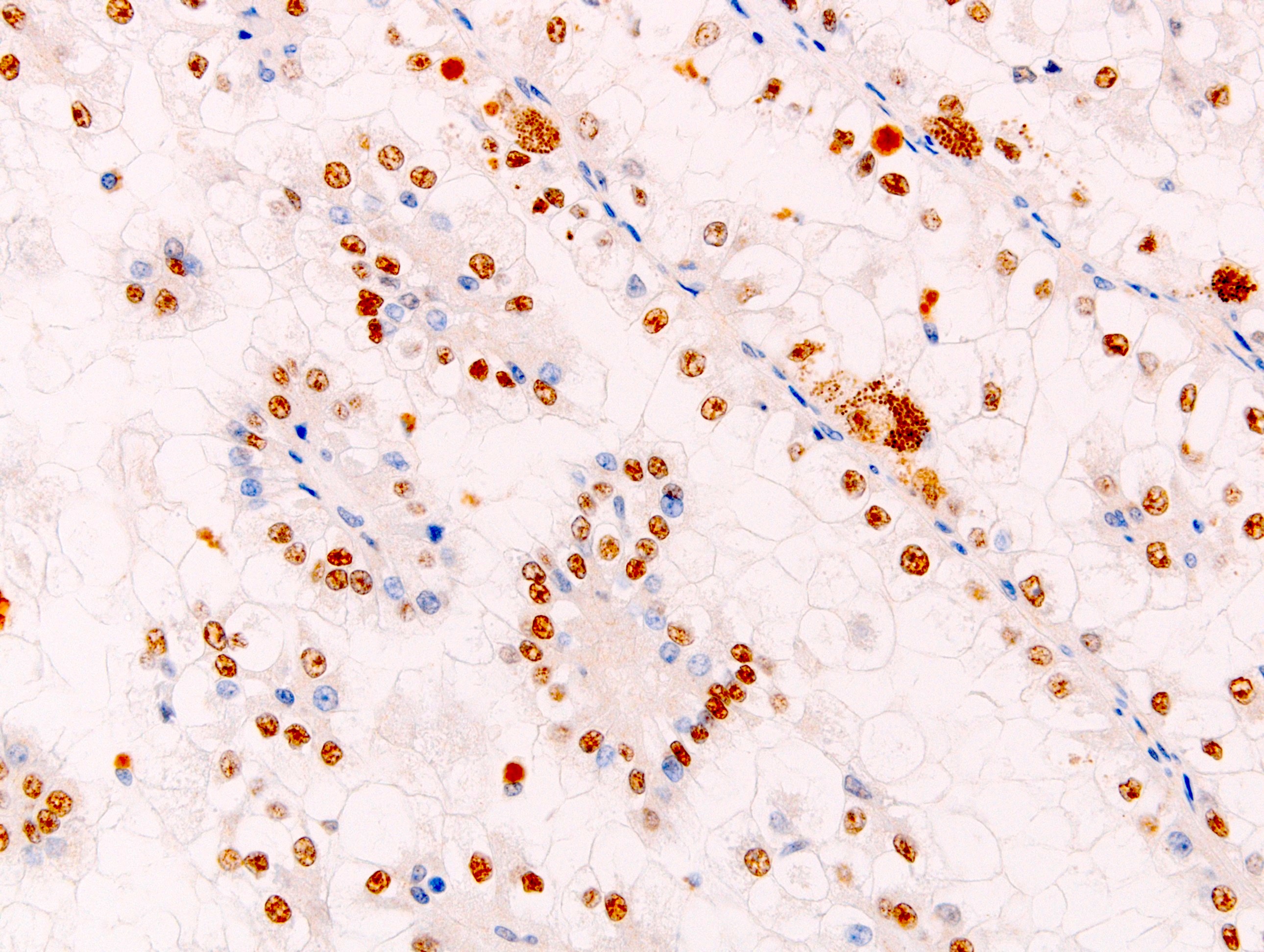
- Diffuse strong nuclear expression in the neoplastic cells
Uses by pathologists
- Differentiate MiT family translocation associated renal cell carcinoma (RCC) from other types of renal cell carcinoma, particularly those with clear cell morphology and papillary architecture
- Sensitivity and specificity depends on type of TFE3 antibody (polyclonal versus monoclonal), tissue fixation and staining conditions (Adv Anat Pathol 2022;29:131, Nat Rev Urol 2014;11:465, Am J Surg Pathol 2003;27:750, Hum Pathol 2019;87:65)
- TFE3 immunostaining is invaluable as a sensitive screening tool but lacks specificity and often requires confirmatory FISH studies (Am J Surg Pathol 2003;27:750, Am J Surg Pathol 2013;37:804, Adv Anat Pathol 2022;29:131)
- Very good sensitivity and specificity when compared with confirmatory FISH studies (Am J Surg Pathol 2003;27:750)
- Aids in the diagnosis of alveolar soft part sarcoma and predicts the presence of the translocation by FISH
- Subset of epithelioid hemangioendotheliomas, PEComas and solid pseudopapillary neoplasms of the pancreas may also be positive for TFE3
Prognostic factors
- Children and young adults with MiT family translocation associated renal cell carcinoma generally show a good prognosis
- Older adults with renal cell carcinoma harboring TFE3 rearrangement by FISH tend to show a worse prognosis (Am J Surg Pathol 2012;36:663)
Microscopic (histologic) images
Positive staining - normal
- TFE3 protein is ubiquitously expressed at low levels in a wide variety of tissues, particularly in the genitourinary tract (J Clin Pathol 2006;59:1127, Adv Anat Pathol 2022;29:131)
Positive staining - disease
- MiT family translocation associated renal cell carcinoma
- Granular cell tumor (Hum Pathol 2014;45:1039)
- Alveolar soft part sarcoma
- Subset of epithelioid hemangioendotheliomas
- Subset of perivascular endothelial cell tumors (PEComas)
- Some solid pseudopapillary pancreatic neoplasms
- Subset of ovarian sclerosing stromal tumors
- Subset (~9%) of RCCs lacking TFE3 rearrangements and associated with higher nuclear grade, advanced tumor stage and metastatic disease, as well as an unfavorable outcome (Adv Anat Pathol 2022;29:131, Histopathology 2018;73:758, BJU Int 2011;108:E71)
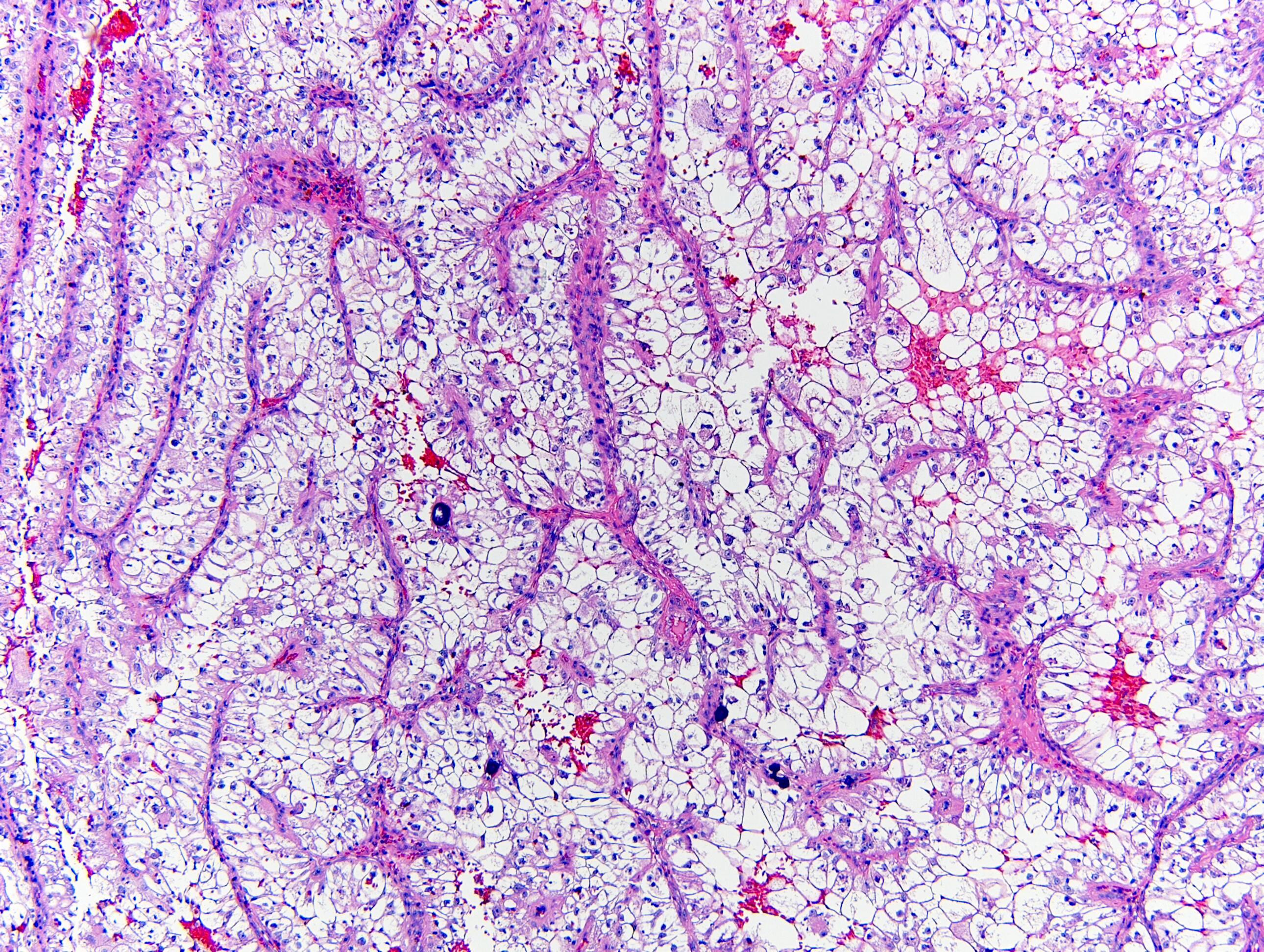
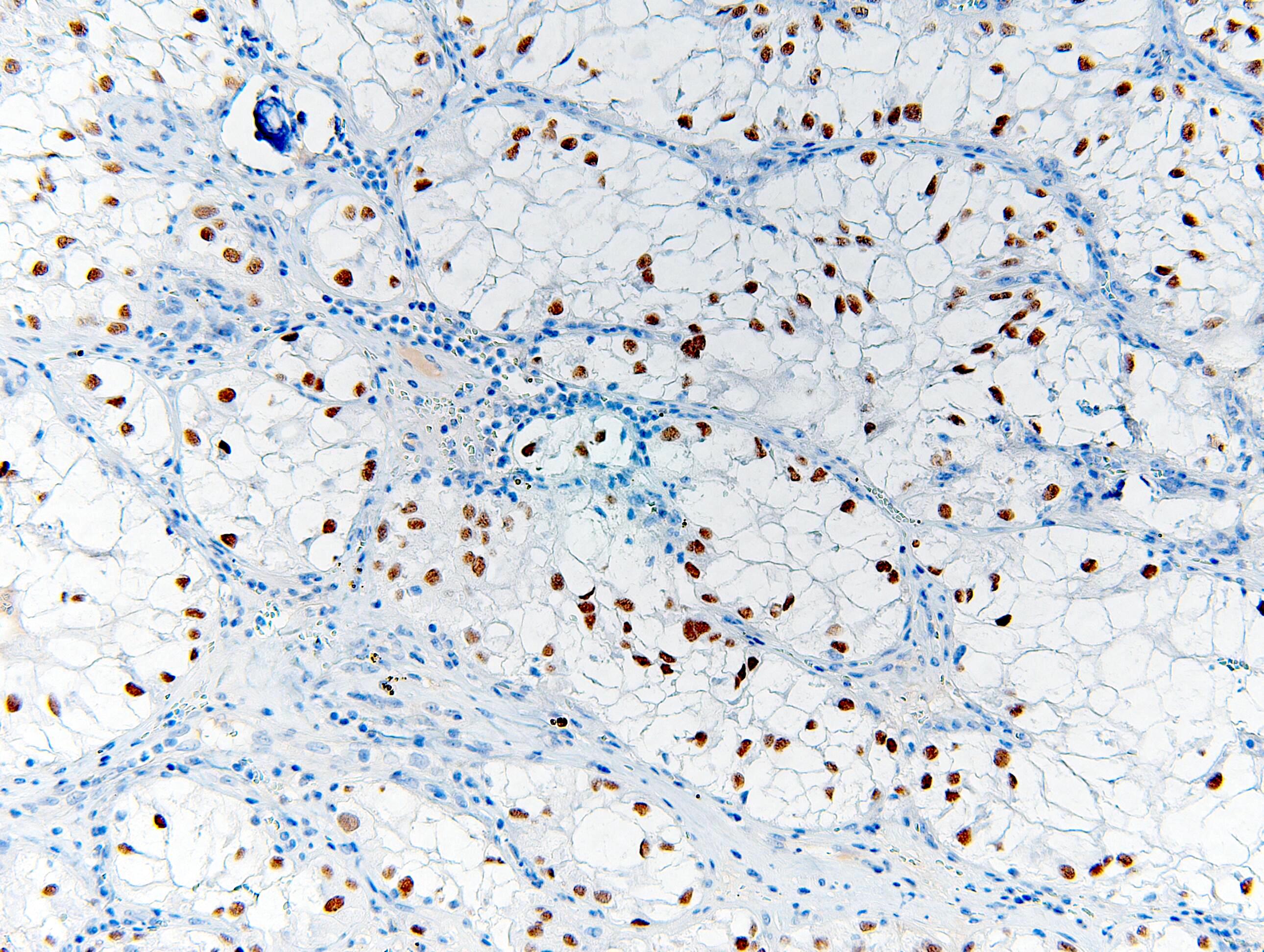
Negative staining
- Clear cell, papillary, clear cell papillary, FH deficient and chromophobe type renal cell carcinoma
- Clear cell sarcoma
- Reference: Am J Surg Pathol 2003;27:750
Molecular / cytogenetics description
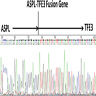
- TFE3 break apart probes in FISH analysis are most effective at detection of the fusion due to multiple partners of the gene
- Most common partner genes in MiT family translocation associated renal cell carcinoma are PRCC and
ASPSCR1 (ASPL) (Am J Surg Pathol 2016;40:723)
- Melanotic subtype generally harbors the SFPQ-TFE3 fusion and rarely ARID1B-TFE3 (Am J Surg Pathol 2017;41:1576)
- SFPQ and NONO are also common partners in certain cases of PEComas (Am J Surg Pathol 2015;39:1181, Histopathology 2016;69:450, Am J Surg Pathol 2017;41:717)
- Fusion most commonly seen in alveolar soft part sarcoma is ASPSCR1 (ASPL) (Oncogene 2001;20:48)
- Epithelioid hemangioendotheliomas often show WWTR1-CAMTA1 fusions but a subset can also show concomitant YAP1-TFE3 (Oncotarget 2016;7:7480)
- Reverse transcriptase polymerase chain reaction (PCR) studies can be performed for a more sensitive detection of the fusion product
- More recently, next generation sequencing using mapping based data mining of sequencing reads can also detect fusions (Am J Surg Pathol 2017;41:1576)
- Some studies have shown RNA sequencing to be an effective method for novel gene fusion detection (Mod Pathol 2018;31:1346)
- While granular cell tumor expresses TFE3 by immunohistochemistry, it does not show TFE3 gene rearrangements by FISH (Hum Pathol 2015;46:1242)
Molecular / cytogenetics images
Sample pathology report
- Right kidney, total nephrectomy:
- MiT family translocation renal cell carcinoma, TFE3 positive
Board review style question #1
Board review style answer #1
D. Melanotic perivascular endothelial cell tumor. TFE3 fusions are most closely associated with Xp11 translocation renal cell carcinoma, alveolar soft part sarcoma, melanotic Xp11 renal cell carcinoma, some PEComas and others. Although increased TFE3 expression is seen in some solid pseudopapillary pancreatic neoplasms, these have been negative for the gene fusion.
Comment Here
Reference: TFE3
Comment Here
Reference: TFE3
Board review style question #2
Which of the following is a common, effective method for detection of the TFE3 gene fusion?
- Break apart FISH probe
- Conventional karyotype analysis
- Dual color, dual fusion FISH probe
- Immunohistochemistry
- RNA sequencing
Board review style answer #2
A. Break apart FISH probe. Due to the multiple translocation partners seen in TFE3 associated neoplasms, a break apart FISH probe is recommended over a dual color, dual fusion FISH probe. Some fusions are cryptic and cannot be detected by conventional karyotype analysis. Immunohistochemistry would detect the overexpression of TFE3 but cannot confirm whether the overexpression is due to the fusion event or other, possibly epigenetic, factors leading to TFE3 overexpression. While RNA sequencing can detect the multiple fusion partners, it is not commonly utilized at this time.
Comment Here
Reference: TFE3
Comment Here
Reference: TFE3