Table of Contents
Definition / general | Acral fibromyxoma | Alveolar soft part sarcoma | Aneurysmal bone cyst, primary | Angiofibroma of soft tissue | Angiomatoid fibrous histiocytoma | Angiosarcoma, radiation associated | Cellular angiofibroma | Chondroblastoma | Chondroid lipoma of soft tissue | Chondromyxoid fibroma | Chondrosarcoma, myxoid, extraskeletal (EMC) | Chordoma, poorly differentiated | CIC rearranged sarcoma | Clear cell sarcoma | Dermatofibrosarcoma protuberans / giant cell fibroblastoma | Desmoid type fibromatosis | Desmoplastic fibroblastoma | Desmoplastic small round cell tumor | Endometrial stromal sarcoma | Epithelioid hemangioendothelioma | Epithelioid hemangioma | Epithelioid sarcoma | Ewing sarcoma / PNET, extraosseous | EWSR1::SMAD3 positive fibroblastic tumor | Fibrous hamartoma of infancy | Giant cell fibroblastoma | Giant cell tumor of bone | Glomus tumor | Infantile fibrosarcoma / mesoblastic nephroma | Inflammatory myofibroblastic tumor | Lipoma | Lipoma, spindle cell / pleomorphic | Liposarcoma, myxoid / round cell variants | Liposarcoma, well differentiated / dedifferentiated | Low grade fibromyxoid sarcoma / sclerosing epithelioid fibrosarcoma | Low grade osteosarcoma | Malignant melanotic nerve sheath tumor | Malignant peripheral nerve sheath tumor | Mesenchymal chondrosarcoma | Myoepithelioma of soft tissue | Myopericytoma, including myofibroma | Myositis ossificans and fibro-osseous pseudotumor of digits | Myxoinflammatory fibroblastic tumor | Neuromuscular choristoma | Nodular fasciitis | NTRK rearranged spindle cell neoplasm | Ossifying fibromyxoid tumor | Osteoid osteoma / osteoblastoma | Phosphaturic mesenchymal tumor | Pseudomyogenic hemangioendothelioma | Rhabdomyosarcoma, alveolar | Round cell sarcoma with EWSR1-non-ETS fusions | Sarcoma with BCOR genetic alterations | Sclerosing epithelioid fibrosarcoma | Spindle cell / sclerosing rhabdomyosarcoma | Superficial CD34 positive fibroblastic tumor | Synovial chondromatosis | Synovial sarcoma | Tenosynovial giant cell tumor | Board review style question #1 | Board review style answer #1 | Board review style question #2 | Board review style answer #2Cite this page: Alexiev BA, Mantilla JG. Soft tissue and bone molecular. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/softtissuemolecular.html. Accessed April 2nd, 2025.
Definition / general
- This topic contains characteristic molecular alterations for topics in our Soft tissue and Bone & joints chapters
- The intent is not to catalog every alteration identified, just those that are considered characteristic
- More information is listed in the linked topics
- Most sarcomas are cytogenetically complex neoplasms
- Recurrent genetic alterations are seen in a subset of bone and soft tissue neoplasms
- Point mutations: detection may aid in diagnosis or allow targeted therapy
- Chromosomal rearrangements: recurrent gene fusions can be useful for diagnosis
- Copy number alterations
Acral fibromyxoma
- Tumor shows deletion of the RB1 gene (Hum Pathol 2017;60:192)
- See Acral fibromyxoma
Alveolar soft part sarcoma
- t(X;17)(p11.2;q25) - ASPSCR1::TFE3 fusion gene
- Note: ASPSCR1 is also known as ASPL
- See Alveolar soft part sarcoma
- Reference: Liddy Shriver Sarcoma Initiative: Chromosomal Translocations in Sarcomas [Accessed 19 February 2021]
Aneurysmal bone cyst, primary
- t(17;17)(p13;q12)
- Up to 75% have USP6 rearrangements
- See Aneurysmal bone cyst
Angiofibroma of soft tissue
- Detection of NCOA2 gene rearrangements (AHRR::NCOA2 gene fusion) supports the diagnosis of angiofibroma of soft tissue (Genes Chromosomes Cancer 2017;56:750)
- See Angiofibroma of soft tissue
Angiomatoid fibrous histiocytoma
- t(2;22)(q33;q12) - EWSR1 / FUS::CREB1 fusion gene (Genes Chromosomes Cancer 2007;46:1051, Clin Cancer Res 2007;13:7314)
- t(12;22)(q13;q12) - EWSR1::ATF1 (also present in clear cell sarcoma)
- t(12;16)(q13;p11) - FUS::ATF1 fusion gene
- See Angiomatoid fibrous histiocytoma
- Reference: Liddy Shriver Sarcoma Initiative: Chromosomal Translocations in Sarcomas [Accessed 19 February 2021]
Angiosarcoma, radiation associated
- Amplification of MYC is consistently seen in radiation associated angiosarcoma (Genes Chromosomes Cancer 2011;50:25)
- Useful for distinguishing from sporadic tumors and radiation associated atypical vascular lesions
- See Angiosarcoma, radiation associated
Cellular angiofibroma
- Tumor shows deletion of the RB1 gene (Mod Pathol 2011;24:82)
- See Cellular angiofibroma
Chondroblastoma
- H3F3B K36M point mutation has been described in 95% of chondroblastomas (Nat Genet 2013;45:1479)
- Mutation specific immunohistochemical stain for H3.3 K36M is helpful to support this diagnosis (Histopathology 2016;69:121)
- See Chondroblastoma
Chondroid lipoma of soft tissue
- t(11,16)(q13;p12-13) - ZFTA (formerly C11orf95)::MRTFB (formerly MKL2) fusion; also in hibernoma (Genes Chromosomes Cancer 2010;49:810, Histopathology 2013;62:925)
- See Chondroid lipoma of soft tissue
Chondromyxoid fibroma
- GRM1 rearrangements (Genes Chromosomes Cancer 2019;58:88)
- See Chondromyxoid fibroma
Chondrosarcoma, myxoid, extraskeletal (EMC)
- t(9;22)(q22;q12) - EWSR1::NR4A3 fusion gene seen in up to 75% of cases
- Other less common rearrangements include t(9;15)(q22;q21) (TFC12::NR4A3) and t(9;17)(q22;q11) (TAF15::NR4A3)
- See Chondrosarcoma, myxoid, extraskeletal
- Reference: Liddy Shriver Sarcoma Initiative: Chromosomal Translocations in Sarcomas [Accessed 19 February 2021]
Chordoma, poorly differentiated
- Molecularly different from conventional chordoma and characterized for SMARCB1 deletion and subsequent loss of INI1 expression (Genes Chromosomes Cancer 2019;58:804)
- See Chordoma
CIC rearranged sarcoma
- t(4;19)(q35;q13) - CIC::DUX4 fusion gene
- t(X;19)(q13;q13) - CIC::FOXO4 fusion gene
- CIC::NUTM1 (Am J Surg Pathol 2019;43:268)
- See CIC rearranged sarcoma
- Reference: Int J Mol Sci 2018;19:3784, Genes Chromosomes Cancer 2012;51:207, Hum Mol Genet 2006;15:2125
Clear cell sarcoma
- t(12;22)(q13;q12) - EWSR1::ATF1 fusion gene
- See Clear cell sarcoma
- Reference: Int J Mol Sci 2018;19:3784
Dermatofibrosarcoma protuberans / giant cell fibroblastoma
- t(17;22)(q22;q13) - COL1A1::PDGFB fusion gene
- COL6A3::PDGFD and EMILIN2::PDGFD (Mod Pathol 2018;31:1683)
- See Dermatofibrosarcoma protuberans / giant cell fibroblastoma
- Reference: Liddy Shriver Sarcoma Initiative: Chromosomal Translocations in Sarcomas [Accessed 19 February 2021]
Desmoid type fibromatosis
- CTNNB1: point mutations in approximately 90% of cases (Virchows Arch 2015;467:203)
- Cases with the S45F mutation may be associated with higher risk of recurrence, compared with wild type and other mutations (Future Oncol 2021;17:435)
- Germline mutations in APC in patients with familial adenomatous polyposis (FAP) (Eur J Surg Oncol 2009;35:3)
- See Desmoid type fibromatosis
Desmoplastic fibroblastoma
- Occasionally t(2;11)(q31;q12) or 11q12 abnormalities
- See Desmoplastic fibroblastoma
Desmoplastic small round cell tumor
- t(11;22)(p13;q12) - EWSR1::WT1 fusion gene
- See Desmoplastic small round cell tumor
- Reference: Int J Mol Sci 2018;19:3784
Endometrial stromal sarcoma
- Low grade endometrial stromal sarcoma: most common rearrangement is t(7;17)(p15;q21) - JAZF1::SUZ12 (JJAZ1)
- However, multiple other fusions, most commonly involving members of the polycomb repressive complex (PRC), have been reported
- High grade endometrial stromal sarcoma: most common rearrangement is t(10;17)(q22;p13), resulting in YWHAE::NUTM2A/B (formerly known as FAM22A/B) (Am J Surg Pathol 2012;36:641)
- ZC3H7B::BCOR, BCOR ITD, JAZF1::PHF1 (Genes Chromosomes Cancer 2013;52:610, Am J Surg Pathol 2018;42:335, PLoS One 2012;7:e39354)
- See Endometrial stromal sarcoma
- Reference: Liddy Shriver Sarcoma Initiative: Chromosomal Translocations in Sarcomas [Accessed 19 February 2021]
Epithelioid hemangioendothelioma
- WWTR1 (TAZ)::CAMTA1 and YAP1::TFE3 (Sci Transl Med 2011;3:98ra82, Genes Chromosomes Cancer 2011;50:644, Genes Chromosomes Cancer 2013;52:775, Mod Pathol 2021;34:2211)
- See Epithelioid hemangioendothelioma
Epithelioid hemangioma
- FOSB gene rearrangements (Am J Surg Pathol 2015;39:1313, Genes Chromosomes Cancer 2014;53:951)
- See Epithelioid hemangioma
Epithelioid sarcoma
- Deletions of SMARCB1 are seen in the vast majority of cases (Adv Anat Pathol 2014;21:394)
- Immunohistochemical loss of nuclear INI1 expression is important for supporting this diagnosis
- See Epithelioid sarcoma
Ewing sarcoma / PNET, extraosseous
- t(11;22)(q24;q12) - EWSR1::FLI1 fusion gene
- t(2;22)(q33;q12) - EWSR1::FEV fusion gene
- t(7,22)(p22;q12) - EWSR1::ETV1 fusion gene
- t(17;22)(q12;q12) - EWSR1::ETV4 fusion gene
- t(21;22)(q22;q12) - EWSR1::ERG fusion gene
- t(16;21)(p11;q22) - FUS::ERG fusion gene
- See Ewing sarcoma / PNET, extraosseous
- Reference: Int J Mol Sci 2018;19:3784
EWSR1::SMAD3 positive fibroblastic tumor
Fibrous hamartoma of infancy
- EGFR exon 20 insertion / duplication (Am J Surg Pathol 2016;40:1713)
- See Fibrous hamartoma of infancy
Giant cell fibroblastoma
- COL1A1::PDFGB fusion may be detected by various molecular approaches (Genes Chromosomes Cancer 2008;47:260)
- See Giant cell fibroblastoma
Giant cell tumor of bone
- Point mutations in p.Gly34 of H3F3A have been reported in 92% of cases (Nat Genet 2013;45:1479)
- G34W substitution in 90% of tumors
- Mutation specific immunohistochemical stain for H3.3 G34W is helpful to support the diagnosis of giant cell tumor (Am J Surg Pathol 2017;41:1059)
- See Giant cell tumor of bone
Glomus tumor
- MIR143::NOTCH fusions or BRAF mutations in benign and malignant glomus tumors (Am J Surg Pathol 2017;41:1532)
- See Glomus tumor
Infantile fibrosarcoma / mesoblastic nephroma
- t(12;15)(p13;q25) - ETV6::NTRK3 fusion gene
- See Infantile fibrosarcoma / mesoblastic nephroma
Inflammatory myofibroblastic tumor
- Translocations at 2p23 involving ALK gene
- t(1;2)(q21;p23) - TMP3::ALK fusion gene
- t(2;19)(p23;p13) - TMP4::ALK fusion gene
- t(2;17)(p23;q23) - CLTC::ALK fusion gene
- ROS1 and NTRK3 rearrangements (Cancer Discov 2014;4:889, Mod Pathol 2015;28:732, Am J Surg Pathol 2016;40:1051)
- See Inflammatory myofibroblastic tumor
- Reference: Int J Mol Sci 2018;19:3784
Lipoma
- t(12;14)(q13-15;q23-24) or related changes involving HMGA2 / HMGIC at 12q13-15
- See Lipoma
Lipoma, spindle cell / pleomorphic
- Deletion in 13q has been described in most cases of pleomorphic / spindle cell lipoma (Genes Chromosomes Cancer 2011;50:619)
- Loss of RB1 (13q14) can be detected by FISH or IHC to support this diagnosis
- This alteration is shared by a spectrum of similar neoplasms, including mammary type myofibroblastoma and cellular angiofibroma (Am J Surg Pathol 2012;36:1119)
- Atypical spindle cell / pleomorphic lipomatous tumor (similar alterations) (Am J Surg Pathol 2017;41:234, Am J Surg Pathol 2017;41:1443)
- See Lipoma, spindle cell / pleomorphic
Liposarcoma, myxoid / round cell variants
- t(12;16)(q13;p11) or t(12;22)(q13;q12) - FUS / EWSR1::DDIT3 (formerly CHOP) fusion gene
- See Liposarcoma, myxoid / round cell variants
- Reference: Int J Mol Sci 2018;19:3784
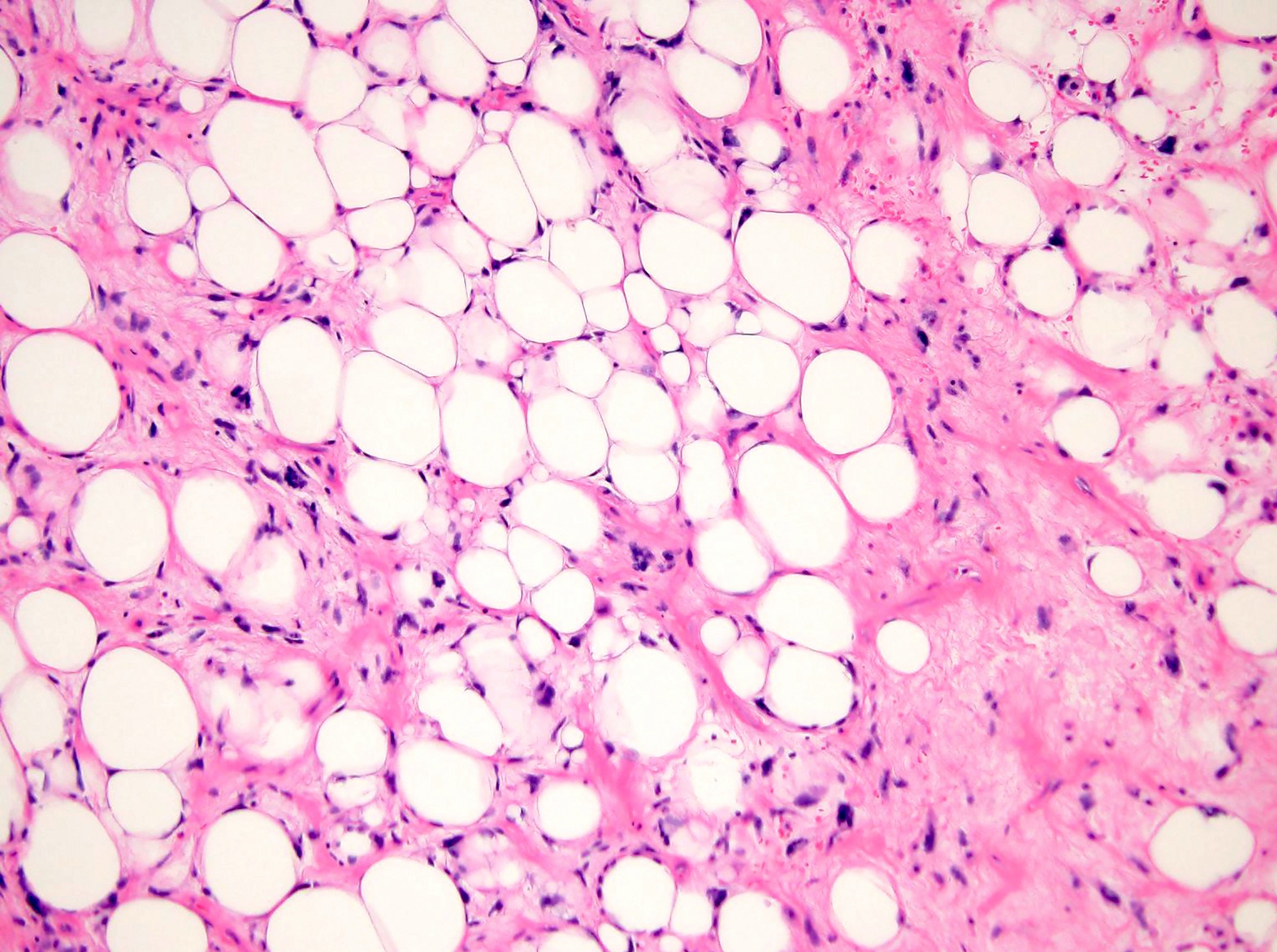
Liposarcoma, well differentiated / dedifferentiated
- Marker ring or giant chromosomes derived from 12q13-15; amplification of MDM2 and CDK4 is characteristic
- Detection of MDM2 amplification by FISH (or IHC expression) is useful for diagnosis
- Potential targeted therapy with CDK4 inhibitors (Crit Rev Oncol Hematol 2020;153:103029)
- See Liposarcoma, well differentiated / dedifferentiated
Low grade fibromyxoid sarcoma / sclerosing epithelioid fibrosarcoma
- t(7;16)(q32-34;p11) - FUS::CREB3L2 or t(11;16)(p11;p11) - FUS::CREB3L1
- See Low grade fibromyxoid sarcoma / sclerosing epithelioid fibrosarcoma
Low grade osteosarcoma
- Consistent amplification of MDM2 gene in low grade osteosarcoma and dedifferentiated low grade osteosarcoma (Am J Surg Pathol 2018;42:1143)
- See Low grade osteosarcoma
Malignant melanotic nerve sheath tumor
- Formerly termed melanotic schwannoma / psammomatous melanotic schwannoma
- PRKAR1A inactivating mutation, also seen in Carney complex (Genes Chromosomes Cancer 2015;54:463, Am J Surg Pathol 2014;38:94)
- See Malignant melanotic nerve sheath tumor
Malignant peripheral nerve sheath tumor
- Mutations involving BRAF V600, which can be used for targeted treatment, seen in a subset of cases (Histopathology 2013;62:499, J Natl Compr Canc Netw 2018;16:967, J Neurooncol 2014;120:267)
- Up to 50% of epithelioid MPNST harbor SMARCB1 deletion
- Inactivating mutations of NF1, CDKN2A / CDKN2B and PRC2 components (EED, SUZ12) and associated H3K27me3 loss (Nat Genet 2014;46:1227, Nat Genet 2014;46:1170, Am J Surg Pathol 2016;40:479)
- See Malignant peripheral nerve sheath tumor
Mesenchymal chondrosarcoma
- HEY1::NCOA2 fusion - reported in up to 80% of cases (Pathol Int 2012;62:823)
- Minority of cases may have IRF2BP2::CDX1 fusion; t(1;5)(q42;q32) (PLoS One 2012;7:e49705)
- See Mesenchymal chondrosarcoma
Myoepithelioma of soft tissue
- EWSR1 is most commonly rearranged, followed by FUS (Genes Chromosomes Cancer 2010;49:1114, Genes Chromosomes Cancer 2015;54:267, Genes Chromosomes Cancer 2020;59:348)
- Fusion partners include ATF1, KLF15, KLF17, PBX1, PBX3, POU5F1, VGLL1 and ZNF444 (Hum Pathol 2012;43:764, Am J Surg Pathol 2016;40:386, Genes Chromosomes Cancer 2008;47:558, Genes Chromosomes Cancer 2015;54:63, Genes Chromosomes Cancer 2010;49:1114, Histopathology 2014;65:917, Genes Chromosomes Cancer 2020;59:249, Genes Chromosomes Cancer 2009;48:1051)
- EWSR1::POU5F1 fusions are associated with malignant myoepithelial tumor with nested epithelioid growth and clear cell morphology (Genes Chromosomes Cancer 2020;59:348)
- Myoepithelial tumors with EWSR1::PBX1/3 fusions are often benign and associated with bland spindle cell and sclerotic morphology (Genes Chromosomes Cancer 2020;59:348)
- FUS::KLF17 fusions are associated with chordoma-like morphology (Genes Chromosomes Cancer 2020;59:348)
- Superficial tumors and those with ductal differentiation tend to genetically approximate mixed tumors / pleomorphic adenomas of salivary gland with PLAG1 rearrangement (Genes Chromosomes Cancer 2013;52:675)
- See Myoepithelioma / myoepithelial carcinoma / mixed tumor
Myopericytoma, including myofibroma
- Recurrent SRF::RELA fusion in a subset of cellular myofibroma / myopericytoma (Am J Surg Pathol 2017;41:677)
- See Myopericytoma / myofibroma
Myositis ossificans and fibro-osseous pseudotumor of digits
- Tumor shows rearrangements of USP6 gene (Int J Surg Pathol 2020;28:816)
- See Myositis ossificans / fibro-osseous pseudotumor of digits
Myxoinflammatory fibroblastic tumor
- t(1;10) (TGFBR3::MGEA5) (Cancer Genet Cytogenet 2008;182:140, J Pathol 2009;217:716)
- BRAF fusions (Am J Surg Pathol 2017;41:1456)
- YAP1::MAML2 in nodular necrotizing variant (Mod Pathol 2022;35:1398)
- VGLL3 amplification (J Pathol 2009;217:716)
- See Myxoinflammatory fibroblastic tumor
Neuromuscular choristoma
- CTNNB1 exon 3 mutations (Am J Surg Pathol 2016;40:1368)
- See Neuromuscular choristoma
Nodular fasciitis
- Gene rearrangements involving USP6 (17p13) are seen in most cases (Mod Pathol 2017;30:1577)
- Various fusion partners, including MYH9, COL1A1, CTNNB1, among many others
- 2 reported cases of malignant transformation of nodular fasciitis harbor PPP6R3::USP6 gene fusion and amplification (Genes Chromosomes Cancer 2016;55:640, Pathol Int 2019;69:706)
- Fusion genes shared with other self limiting neoplasms such as aneurysmal bone cyst, cranial fasciitis, cellular fibroma of tendon sheath and fibro-osseous pseudotumor of digits (Mod Pathol 2021;34:13)
- See Nodular fasciitis
NTRK rearranged spindle cell neoplasm
Ossifying fibromyxoid tumor
- PHF1 gene rearrangement has been observed in 80%, including benign, atypical and malignant subtypes, with fusion to EP400 in 44% of cases (Arch Pathol Lab Med 2016;140:371, Hum Pathol 2013;44:2603)
- ZC3H7B::BCOR and MEAF6::PHF1 fusions occur predominantly in S100 negative and malignant OFMT (Genes Chromosomes Cancer 2014;53:183)
- Recurrent PHF1::TFE3 fusions in a subset of OFMTs with aggressive clinical behavior (Genes Chromosomes Cancer 2019;58:643)
- See Ossifying fibromyxoid tumor
Osteoid osteoma / osteoblastoma
- FOS / FOSB rearrangements (Genes Chromosomes Cancer 2019;58:88)
- See Osteoid osteoma / osteoblastoma
Phosphaturic mesenchymal tumor
- Tumor shows FN1::FGFR1 and FN1::FGF1 fusion genes (Mod Pathol 2016;29:1335)
- See Phosphaturic mesenchymal tumor
Pseudomyogenic hemangioendothelioma
- SERPIN1::FOSB and ACTB::FOSB (J Pathol 2014;232:534, Am J Surg Pathol 2018;42:1653)
- See Pseudomyogenic hemangioendothelioma
Rhabdomyosarcoma, alveolar
- t(2;13)(q35;q14) - PAX3::FOXO1 or t(1;13)(p36;q14) - PAX7::FOXO1
- See Rhabdomyosarcoma, alveolar
- Reference: Int J Mol Sci 2018;19:3784
Round cell sarcoma with EWSR1-non-ETS fusions
- Tumors with non-ETS gene partners were previously designated Ewing-like sarcoma but are now classified as round cell sarcoma with EWSR1-non-ETS fusions in the latest 2020 WHO classification
- There may be significant correlations between fusion subtype and age at presentation, primary tumor sites and overall survival in these patients (Genes Chromosomes Cancer 2020;59:525)
- Includes EWSR1 / FUS::NFATC2 and EWSR1::PATZ1 (Am J Surg Pathol 2019;43:1112, Hum Pathol 2019;90:45, Mod Pathol 2019;32:1593, Am J Surg Pathol 2019;43:220)
- EWSR1::NFATC2 is also seen in simple bone cysts and epithelioid vascular neoplasm of bone (Am J Surg Pathol 2020;44:1623, Genes Chromosomes Cancer 2021;60:762)
Sarcoma with BCOR genetic alterations
- BCOR::CCNB3 sarcoma (BCS) is a recently defined genetic entity among undifferentiated round cell sarcomas, initially classified as and treated similarly to the Ewing sarcoma (ES) family of tumors
- In contrast to ES, BCS shows consistent BCOR overexpression and preliminary evidence suggests that these tumors share morphologic features with other tumors harboring BCOR genetic alterations, including BCOR internal tandem duplication (ITD) and BCOR::MAML3 (Am J Surg Pathol 2018;42:604)
- BCOR - BCL6 transcriptional corepressor is located at Xp11.4 locus (Genes Dev 2000;14:1810, J Clin Pathol 2020;73:314)
- See BCOR
Sclerosing epithelioid fibrosarcoma
- Tumor shows EWSR1::CREB3L1, EWSR1::CREB3L2 and FUS::CREB3L2 gene fusions (Genes Chromosomes Cancer 2020;59:217)
- Recurrent YAP1 and KMT2A gene rearrangements in a subset of MUC4 negative sclerosing epithelioid fibrosarcoma (Am J Surg Pathol 2020;44:368)
- See Sclerosing epithelioid fibrosarcoma
Spindle cell / sclerosing rhabdomyosarcoma
- VGLL2 related gene fusions in an infantile subset of spindle cell rhabdomyosarcoma (Mod Pathol 2019;32:27)
- MYOD1 mutations commonly associated with sclerosing morphology (Mod Pathol 2019;32:27)
- Also NCOA2 gene rearrangements (Genes Chromosomes Cancer 2013;52:538)
- See Spindle cell / sclerosing rhabdomyosarcoma
Superficial CD34 positive fibroblastic tumor
- PRDM10 rearrangements (Am J Surg Pathol 2019;43:504, Clin Cancer Res 2015;21:864)
- See Superficial CD34 positive fibroblastic tumor
Synovial chondromatosis
- FN1::ACVR2A and ACVR2A::FN1 (Genes Chromosomes Cancer 2019;58:88)
- See Synovial chondromatosis
Synovial sarcoma
- t(X;18)(p11.23;q11) - SS18::SSX1 or t(X;18)(p11.21;q11) - SS18::SSX2 or t(X;18)(p11;q11) - SS18::SSX4 fusion genes
- See Synovial sarcoma
- Reference: Int J Mol Sci 2018;19:3784
Tenosynovial giant cell tumor
- t(1;2)(p11;q35-37)
- Also CSF1 rearrangements (Am J Surg Pathol 2007;31:970, Proc Natl Acad Sci U S A 2006;103:690)
- See Tenosynovial giant cell tumor
Board review style question #1
Which of the following is true about atypical lipomatous tumor / well differentiated liposarcoma?
- Absence of MDM2 amplification
- FUS::DDIT3 or EWSR1::DDIT3 fusions are almost always present
- Loss of RB1 expression is observed in 50 - 70% of cases
- MDM2 amplification is almost always present
- Usually harbors USP6 rearrangement
Board review style answer #1
D. MDM2 amplification is almost always present
Comment Here
Reference: Soft tissue and bone molecular
Comment Here
Reference: Soft tissue and bone molecular
Board review style question #2
Board review style answer #2