Table of Contents
Definition / general | Essential features | Terminology | ICD coding | Types of ISH | ISH probes | Specimen types | Applications | Cell enrichment methods | Interpretation of ISH signals | Advantages versus limitations of fluorescence and brightfield ISH | Uses by pathologists | Molecular / cytogenetics images | Sample FISH report | Board review style question #1 | Board review style answer #1 | Board review style question #2 | Board review style answer #2Cite this page: Caramelo A, Polónia A. FISH overview. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/molecularpathfishgeneral.html. Accessed April 2nd, 2025.
Definition / general
- Detection of specific nucleic acid sequences (DNA / RNA)
- Use of a labeled complementary nucleic acid probe
- Visualization in the cell or tissue (in situ) on a slide by fluorescent microscopy
Essential features
- Detection of specific nucleic acid sequences (DNA / RNA) (foreign or native)
- Visualization of signals in the cell or tissue (in situ) by either digital imaging or manual microscopy
- Detection of numerical chromosome abnormalities (congenital or acquired aneuploidies)
- Detection of amplification / deletion / chromosomal translocation
Terminology
- ISH: in situ hybridization
- FISH: fluorescence in situ hybridization
- BISH: brightfield in situ hybridization
- SISH: silver in situ hybridization
- CISH: chromogenic in situ hybridization
- DDISH: dual hapten dual probe in situ hybridization
- PCR: polymerase chain reaction
ICD coding
- ICD-10: Q90 - Q99 - chromosomal abnormalities, not elsewhere classified
Types of ISH
- Fluorescence ISH (FISH): direct detection of a fluorescent labeled probe
- Brightfield ISH (BISH): indirect detection through an enzymatic reaction
- Silver ISH: detection of silver precipitation (black dots)
- Chromogenic ISH (CISH): detection of chromogen (e.g., red)
- Dual hapten dual probe ISH (DDISH): CISH + SISH
- Hybrid techniques
- Specific to allele PCR - FISH (Nat Protoc 2007;2:2782, Nat Methods 2008;5:877)
- Padlock probes with in situ rolling circle amplification (RCA) (Nat Methods 2010;7:395, Science 1994;265:2085)
ISH probes
- Large chromosome sequences (e.g., large regions, short arm, long arm)
- Repetitive sequences (e.g., centromeres)
- Unique sequences (e.g., specific genes, parts of genes)
Specimen types
- Cells in culture
- Paraffin block sections
- Aspirate smears
- Tissue imprints
- Liquid based preparations
Applications
- Interphase cytogenetics (analysis of chromosomes in nondividing cells)
- Detection of numerical chromosome abnormalities (congenital or acquired aneuploidies)
- Detection of gene amplification (e.g., HER2 in breast / gastric / colorectal / endometrial cancer) (Arch Pathol Lab Med 2023;147:993, Arch Pathol Lab Med 2016;140:1345, Lancet Oncol 2016;17:738, Int J Gynecol Pathol 2021;40:17)
- Detection of gene or chromosomal deletion (e.g., TP53 deletion on chromosome 17p in myeloid neoplasms)
- Detection of chromosomal translocation (e.g., ALK translocation on lung cancer) (Arch Pathol Lab Med 2020 May 13 [Epub ahead of print], Arch Pathol Lab Med 2018;142:321)
- Detection of gene translocation and amplification (e.g., MYC)
- Detection of gene translocation (e.g., cyclin D1)
- Detection of gene translocation (e.g., PML::RARA)
- Viral infections: detection of viral genome (e.g., EBV, HPV, HIV, fungal species)
- Gene expression: detection of messenger RNA for various peptides (e.g., immunoglobulin light chains, albumin)
- Single transcript analysis: detection of point mutations, single nucleotide polymorphisms (SNPs), RNA edited transcripts, tissue specific allele expression, alternative splicing
Cell enrichment methods
- Objective: increase sensitivity of ISH for detection of cytogenetic abnormalities
- Technique: based on physical properties (size, deformability, density) and expression of specific surface antigens
- Applications: plasma cells in bone marrow, circulating tumor cells in blood samples (Arch Pathol Lab Med 2013;137:625, PLoS One 2020;15:e0237308, J Hematol Oncol 2019;12:48)
Interpretation of ISH signals
- Presence of signals (e.g., presence of EBER in EBV infection)
- Gain of signals (e.g., ERBB2 amplification in breast / gastric / colorectal / endometrial cancer)
- Breast cancer (Arch Pathol Lab Med 2023;147:993)
- ISH group 1: HER2/CEP17 ratio ≥ 2.0 and average HER2 copy number ≥ 4.0 signals/cell
- ISH group 2: HER2/CEP17 ratio ≥ 2.0 and average HER2 copy number < 4.0 signals/cell
- ISH group 3: HER2/CEP17 ratio < 2.0 and average HER2 copy number ≥ 6.0 signals/cell
- ISH group 4: HER2/CEP17 ratio < 2.0 and average HER2 copy number ≥ 4.0 and < 6.0 signals/cell
- ISH group 5: HER2/CEP17 ratio < 2.0 and average HER2 copy number < 4.0 signals/cell
- Gastric cancer (Arch Pathol Lab Med 2016;140:1345)
- HER2 positive if HER2/CEP17 ratio ≥ 2.0 or HER2/CEP17 < 2.0 and average HER2 copy number ≥ 6.0 signals/cell
- Colorectal cancer (Lancet Oncol 2016;17:738)
- HER2 positive if HER2/CEP17 ratio ≥ 2.0 in ≥ 50% of cells
- Endometrial cancer (Int J Gynecol Pathol 2021;40:17)
- HER2 positive if HER2/CEP17 ratio ≥ 2.0
- Breast cancer (Arch Pathol Lab Med 2023;147:993)
- Loss of signals (e.g., loss of 1p / 19q in oligodendroglioma)
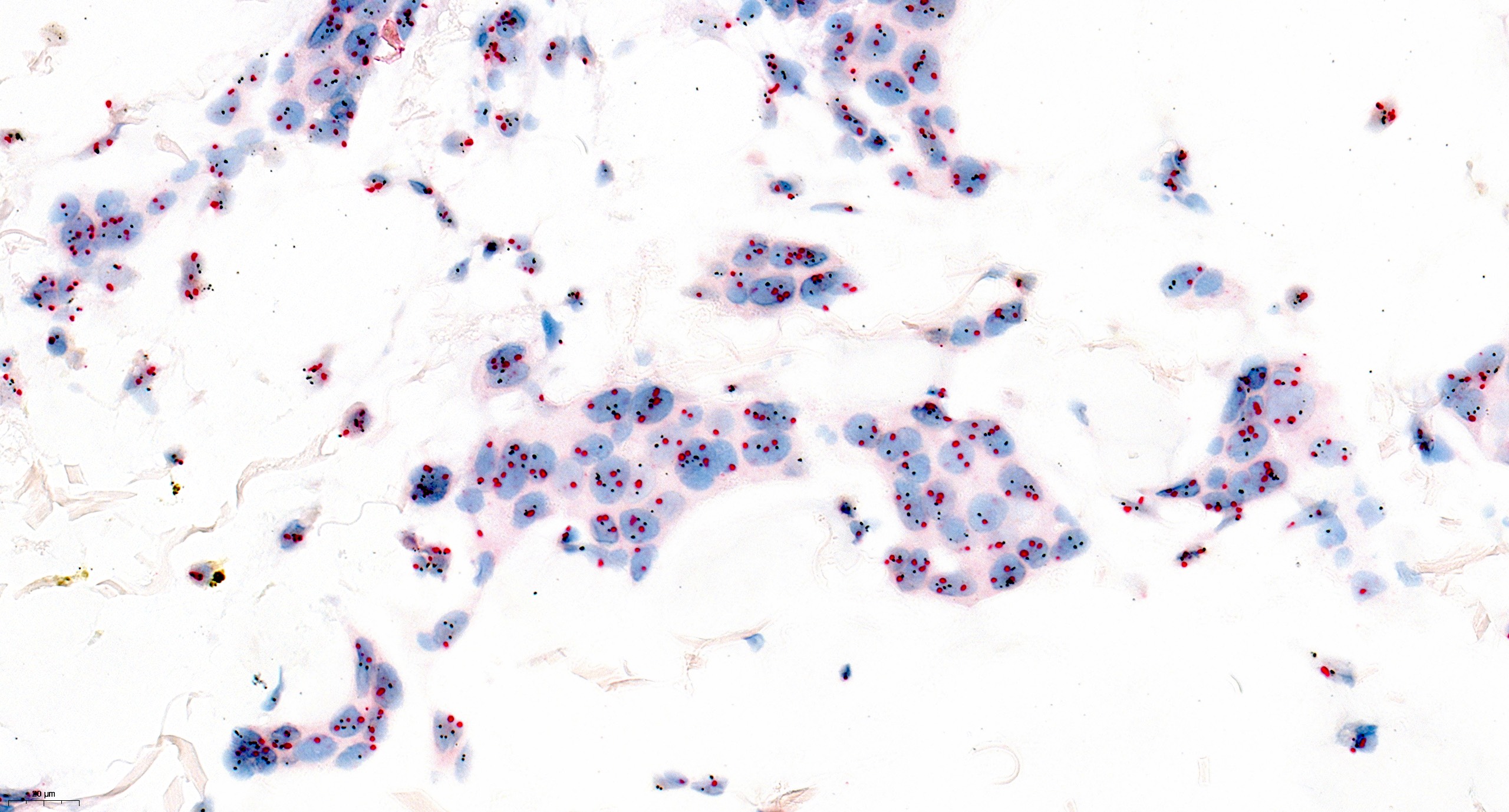
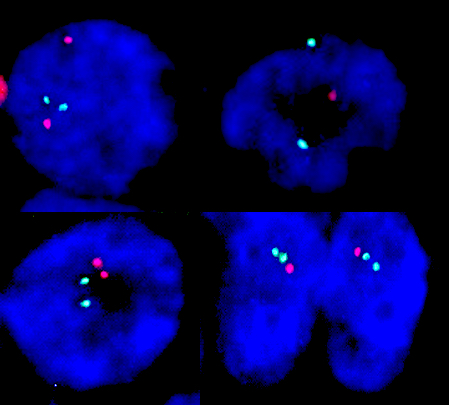
- Positioning of signals
- Break apart probe: neighboring signals altered when detected at a distance, resulting in 2 separate color signals (e.g., EWSR1 translocation)
- Normally, 2 different color signals (red and green) are located nearby, sometimes creating a fused color (yellow); the abnormality exists if the 2 signals are separated
- Fusion probes: distant signals altered when detected nearby, resulting in overlap or juxtaposition of the 2 different color signals (e.g., EWSR1::FLI1 fusion)
- Normally, 2 different color signals (red and green) are separated; the abnormality exists if the 2 signals are together, sometimes creating a fused color (yellow)
- Break apart probe: neighboring signals altered when detected at a distance, resulting in 2 separate color signals (e.g., EWSR1 translocation)
Advantages versus limitations of fluorescence and brightfield ISH
- FISH requires fluorescence microscope (higher cost) versus brightfield ISH requires light microscope (lower cost)
- Higher multiplexing in FISH (> 2 probes) versus low multiplexing in brightfield ISH (≤ 2 probes)
- Low morphologic correlation in FISH versus high morphologic correlation in brightfield ISH
- Loss of signals with time in FISH slides (photographic record mandatory) versus permanent staining in brightfield ISH slides
- Reference: Diagn Pathol 2008;3:41
Uses by pathologists
- See Applications
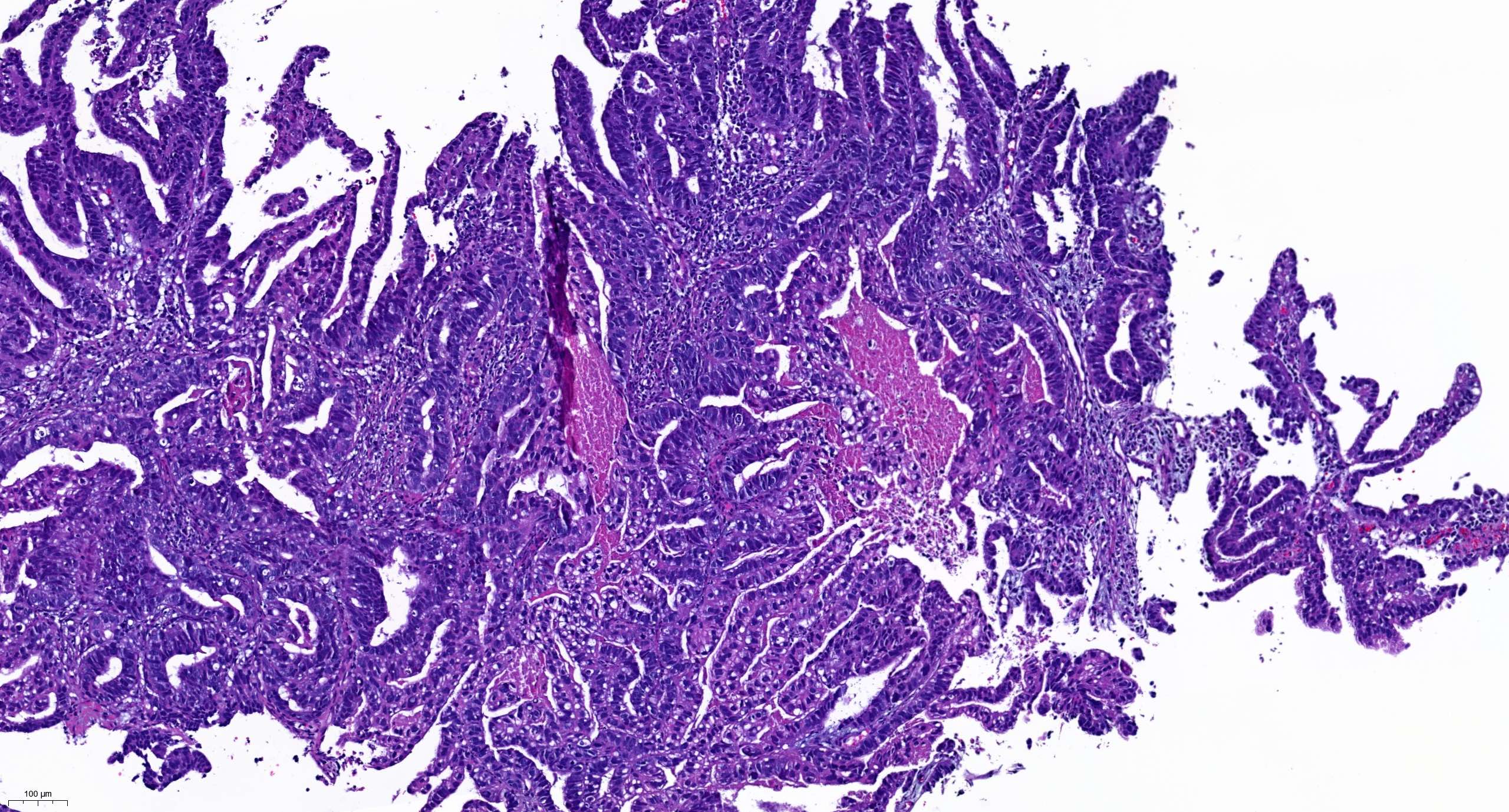
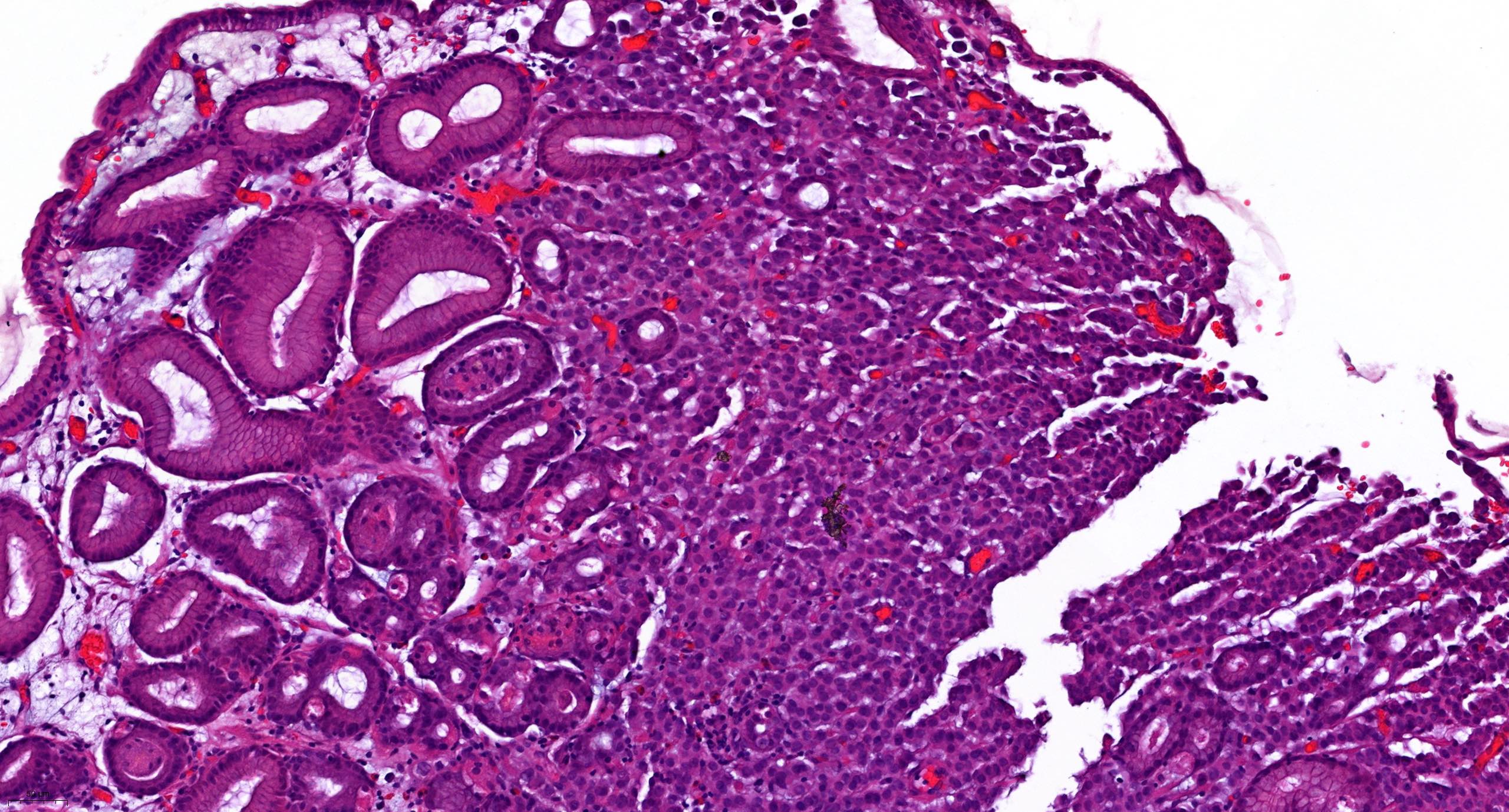
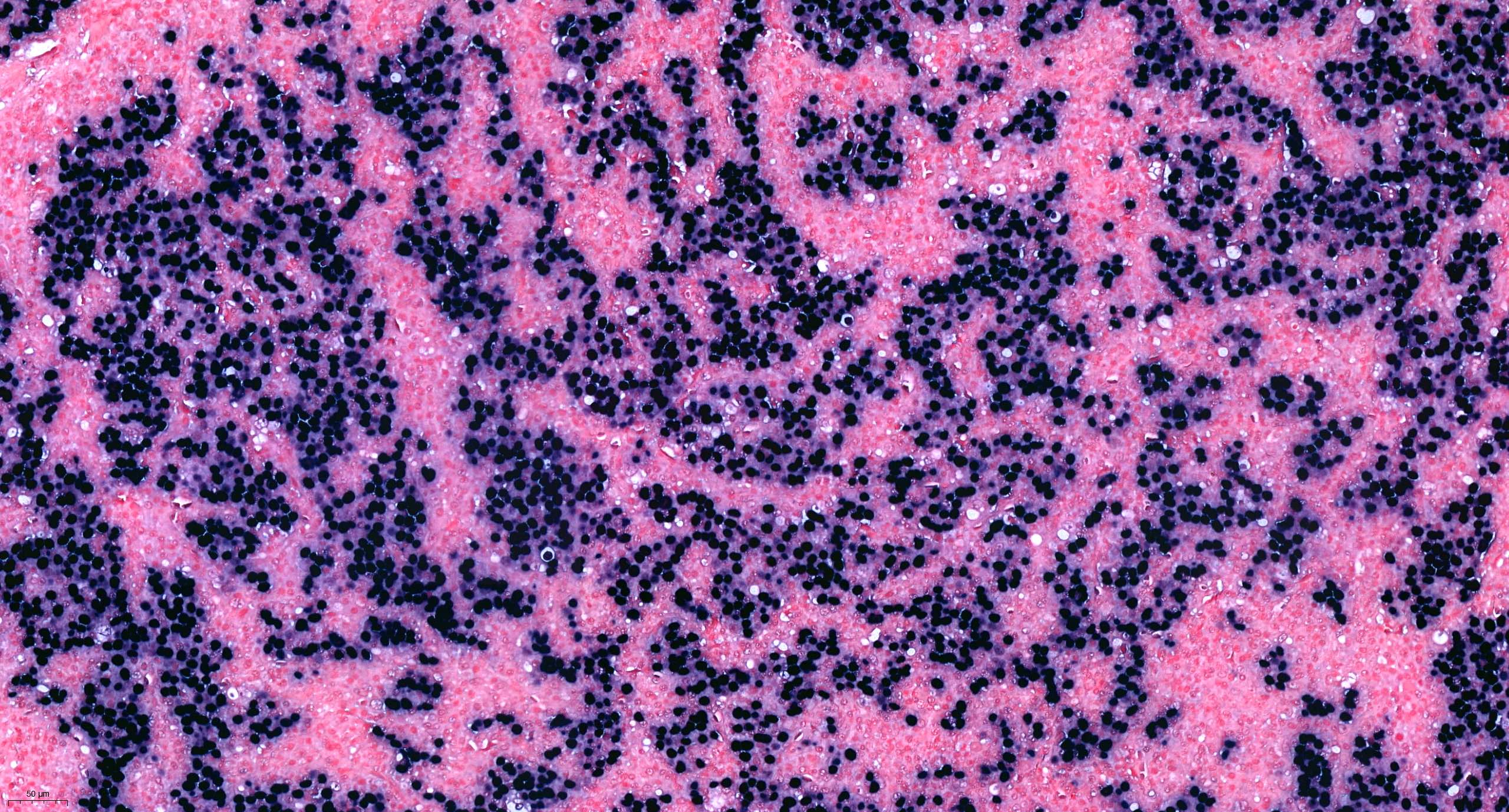
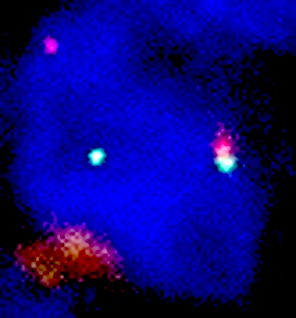
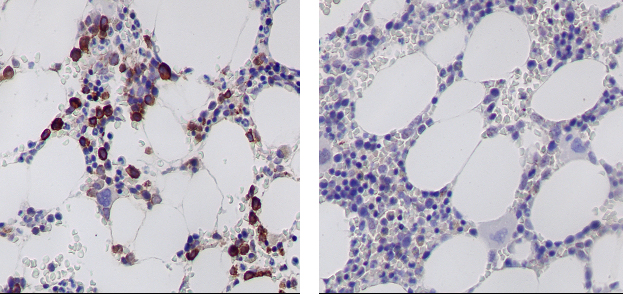
Molecular / cytogenetics images
Contributed by António Polónia, M.D., Ph.D., Ana Caramelo, B.Sc., Ana Ribeiro, M.D.,
Catarina Eloy, M.D., João Vale, M.Sc. and Leica Microsystems, Biosystems division
Images hosted on other servers:
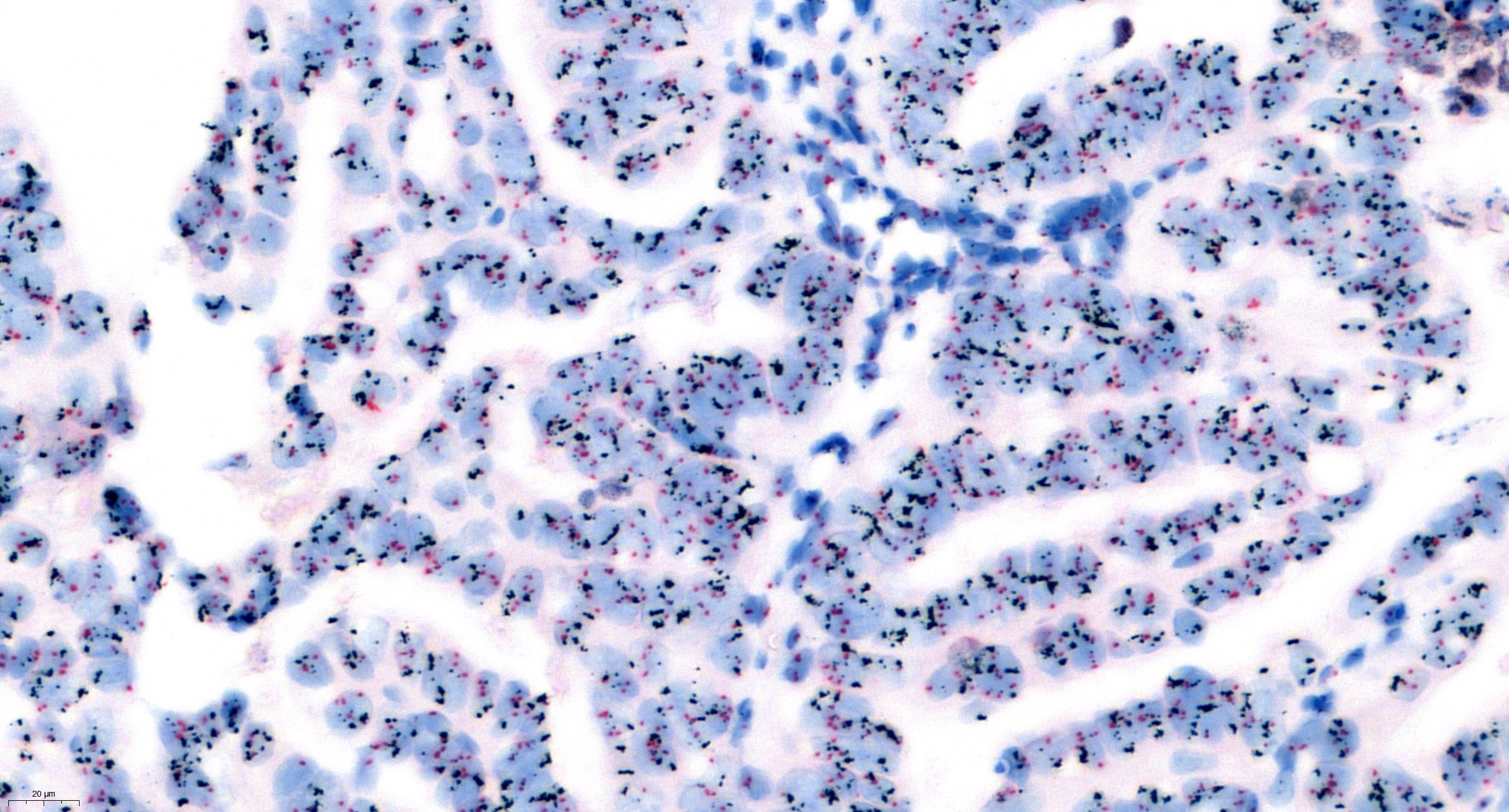
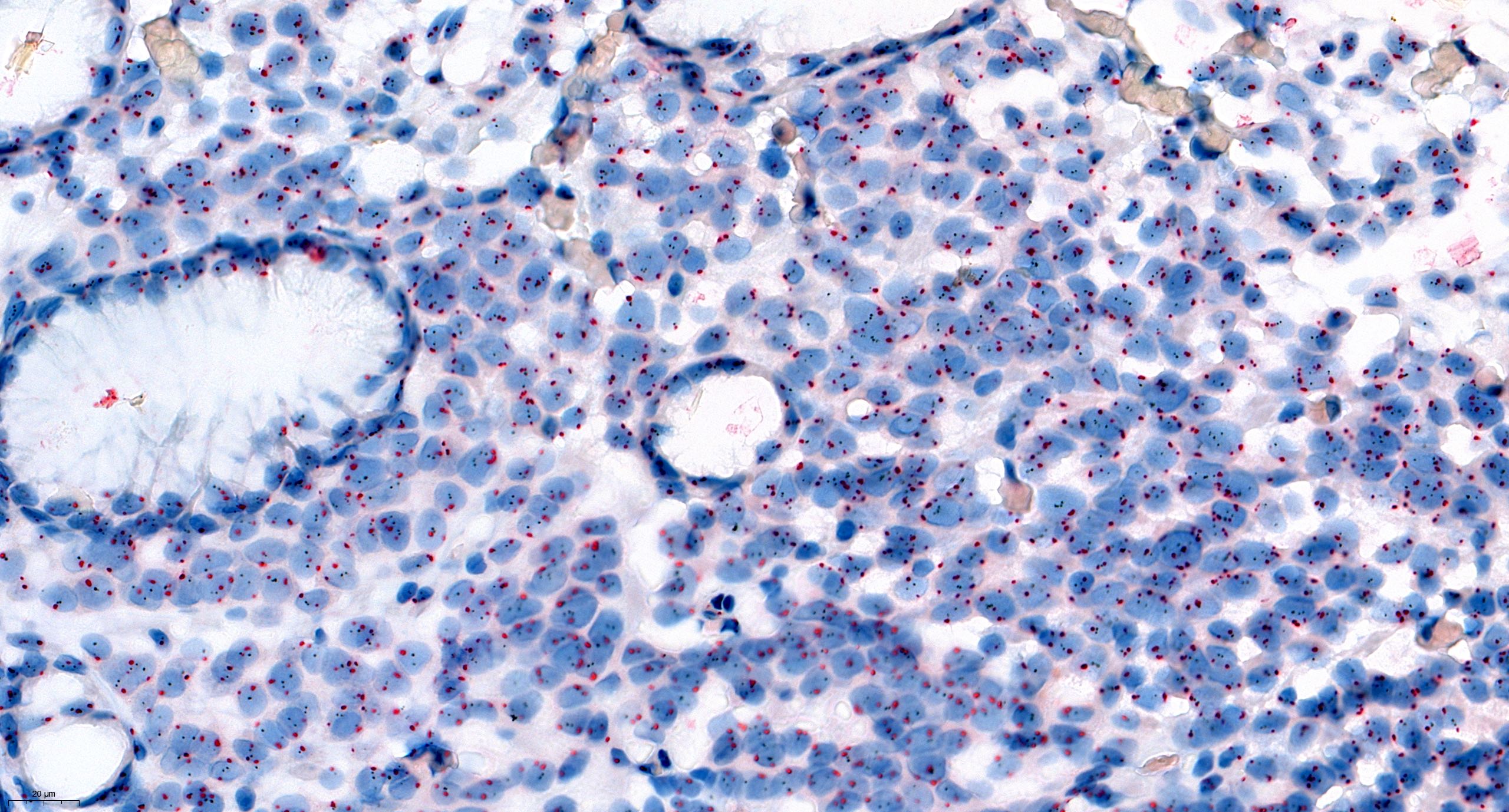
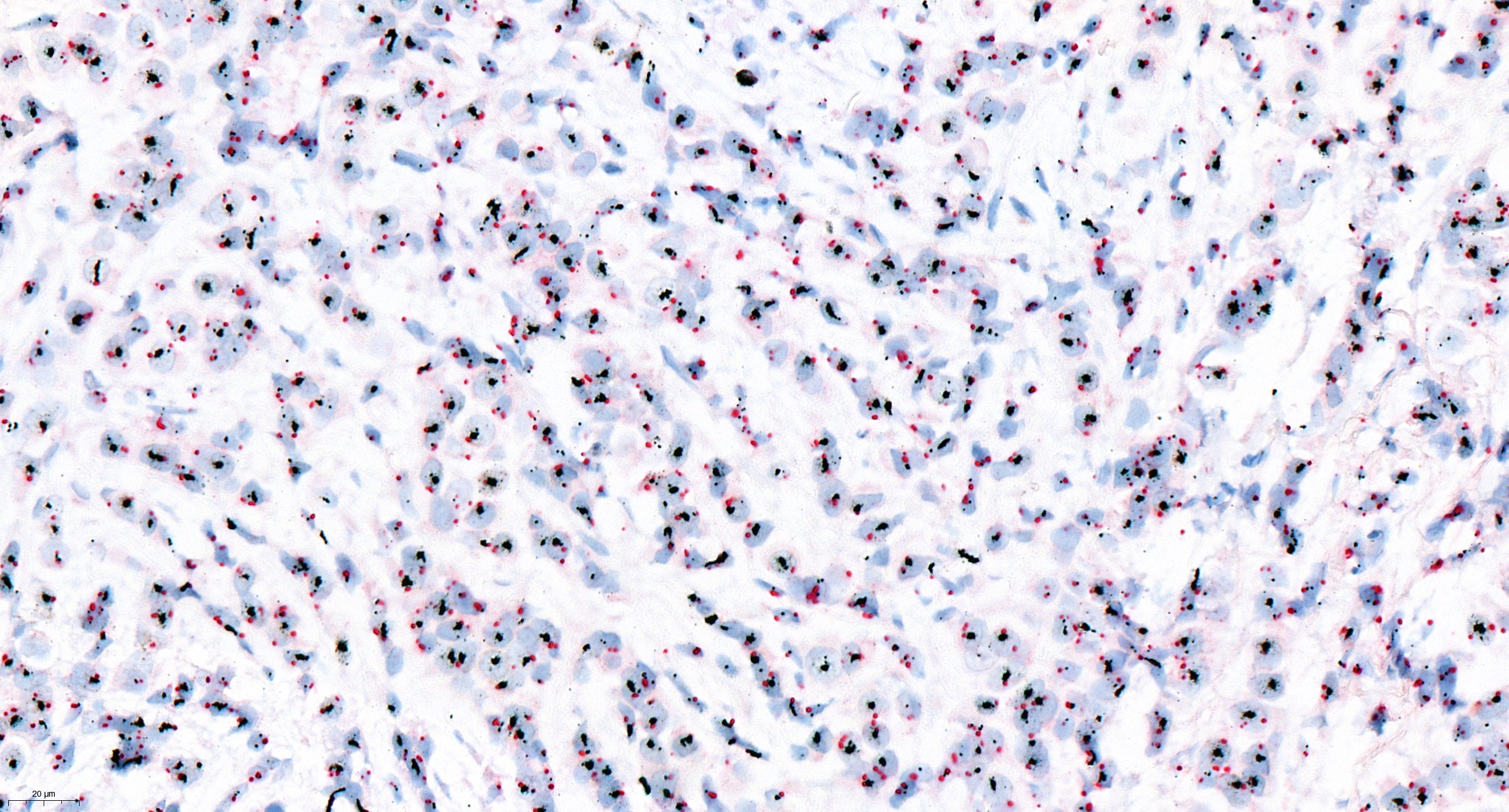
Sample FISH report
- Example for HER2 FISH quantification in breast cancer:
- Number of neoplastic cells analyzed: at least 20 cells in 2 separate areas (at least 10/area)
- Number of HER2 signals: number of black dots
- Average HER2 copy number: number of black dots / number of analyzed cells
- Number of CEP17 signal: number of red dots
- Average CEP17 copy number: number of red dots / number of analyzed cells
- HER2/CEP17 ratio: number of black dots / number of red dots
- Genomic heterogeneity: absence / presence (percentage of the total tumor population with amplification)
- Probe: specify vender's probe
- Guidelines: ASCO / CAP 2023
- Type of fixative: formol / other
- Time to fixation (cold ischemic time): less than 1 hour
- Duration of tissue fixation: between 6 and 72 hours
- Commentary on the preanalytic conditions: the preanalytic conditions are (or are not) according to the recommendations of the ASCO / CAP 2023 guidelines for HER2 in breast cancer.
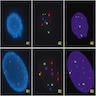
Board review style question #1
Board review style answer #1
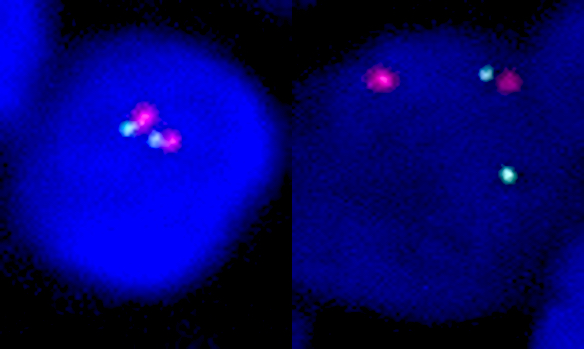
A. 1 pair of different color signals together and 2 separate color signals.
There are 4 signals with 2 different colors. 2 separate color signals (abnormal separate signal) and 2 joint color signals (normal fused signal).
Answer B is incorrect because 2 pairs of different color signals together would represent the normal 2 fused signals.
Answer C is incorrect because 3 pair of different color signals together would represent 3 fused signals.
Answer D is incorrect because 4 different color signals separate would represent 2 pairs of abnormal separate signals.
Comment Here
Reference: FISH overview
Comment Here
Reference: FISH overview
Board review style question #2
What is the pattern of a fusion rearrangement?
- 1 pair of different color signals together and 2 separate color signals
- 2 pairs of different color signals together
- 3 pairs of different color signals together
- 4 different color signals separate
Board review style answer #2
A. 1 pair of different color signals together and 2 separate color signals.
There are 4 signals with 2 different colors. 2 separate color signals (normal separate signal) and 2 joint color signals (abnormal fused signal).
Answer B is incorrect because 2 pairs of different color signals together would represent 2 fused signals.
Answer C is incorrect because 3 pairs of different color signals together would represent 3 fused signals.
Answer D is incorrect because 4 different color signals separate would represent 2 pairs of normal separate signals.
Comment Here
Reference: FISH overview
Comment Here
Reference: FISH overview