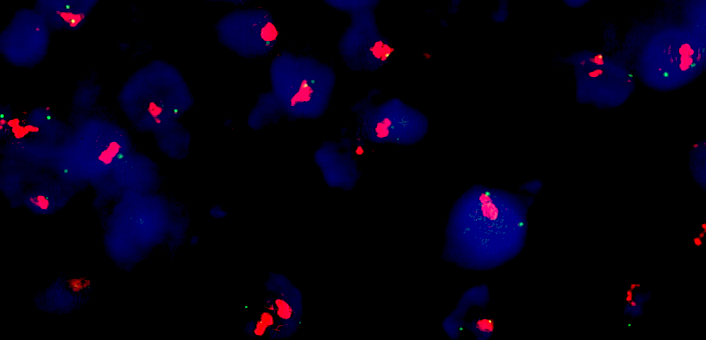Table of Contents
Definition / general | Essential features | Terminology | Pathophysiology | Diagrams / tables | Clinical features | Interpretation | Uses by pathologists | Prognostic factors | Molecular / cytogenetics description | Molecular / cytogenetics images | Sample pathology report | Board review style question #1 | Board review style answer #1Cite this page: Koopman B, van Kempen LC. MET. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/molecularMET.html. Accessed April 3rd, 2025.
Definition / general
- MET proto-oncogene, tyrosine receptor kinase (OMIM: MET Protooncogene; MET [Accessed 30 July 2021])
- Proto-oncogene at 7q31.2; 1390 amino acids with 6.8kb mRNA (OMIM: MET Protooncogene; MET [Accessed 30 July 2021], NCBI: Homo Sapiens MET Proto-oncogene, Receptor Tyrosine Kinase (MET), Transcript Variant 2, mRNA [Accessed 30 July 2021])
- Activates various signaling pathways that lead to proliferation and cell survival (Ther Adv Med Oncol 2011;3:S21)
- Oncogenic activation by amplification, exon 14 skipping mutations and gene fusions (Clin Cancer Res 2016;22:3048)
Essential features
- High level amplification, exon 14 skipping, fusions and kinase domain mutations of MET drive a subset of non small cell lung cancer (NSCLC) and may be targetable with MET inhibition (Clin Cancer Res 2016;22:3048, Nat Med 2016;22:1314, N Engl J Med 2020;383:944, N Engl J Med 2020;383:931, Clin Cancer Res 2018;24:1337)
- MET alterations are targetable resistance mechanisms to EGFR inhibitors and ALK inhibitors (Science 2007;316:1039, Clin Cancer Res 2020;26:2535)
- Amplifications are detected using fluorescent in situ hybridization (FISH) (J Thorac Oncol 2017;12:15, Nat Rev Clin Oncol 2020;17:569)
- Exon 14 skipping mutations are detected with next generation sequencing (NGS) or RNA analysis (Cancer Discov 2015;5:850)
- Immunohistochemistry (IHC) correlates poorly with amplifications and exon 14 skipping and therefore has no current clinical use for pathologists (J Thorac Oncol 2019;14:1666)
Terminology
- C mesenchymal epithelial transition factor (c-Met) (Lancet Oncol 2009;10:709)
- MET proto-oncogene, receptor tyrosine kinase (GeneCards: MET Gene (Protein Coding) [Accessed 30 July 2021])
- Hepatocyte growth factor receptor (HGFR) (GeneCards: MET Gene (Protein Coding) [Accessed 30 July 2021])
- Scatter factor receptor (SFR) (GeneCards: MET Gene (Protein Coding) [Accessed 30 July 2021])
Pathophysiology
- Biology:
- Ligands: hepatocyte growth factor / scatter factor (HGF / SF) and its isoforms (NK1, NK2) (Mol Cell Biol 1998;18:1275, Mol Cell Biol 2000;20:2055)
- Tyrosine receptor kinase that activates the PI3K, MAPK, SRC and STAT signal transduction pathways (Ther Adv Med Oncol 2011;3:S21)
- Involved in embryonic development, wound healing, organ regeneration and tissue remodeling (OMIM: MET Protooncogene; MET [Accessed 30 July 2021])
- Regulates proliferation, scattering, morphogenesis and survival (OMIM: MET Protooncogene; MET [Accessed 30 July 2021])
- Amplification:
- Copy number gain of the MET gene due to focal / regional gene duplication (J Thorac Oncol 2017;12:15)
- Divided into high level or low level amplification and polysomy
- Defined by amount of MET gene copies per cell or ratio between MET gene and chromosome 7 copies (J Thorac Oncol 2017;12:15, Nat Rev Clin Oncol 2020;17:569)
- Cut off values are not internationally standardized (J Thorac Oncol 2017;12:15, Nat Rev Clin Oncol 2020;17:569)
- Exon 14 skipping mutations:
- Most frequent type of MET oncogenic alterations
- Result in an exon 13 - exon 15 fused mRNA transcript, skipping the juxtamembrane domain on exon 14 that binds CBL, resulting in oncogenic activity due to loss of downregulation of signaling activity (Cancer Res 2006;66:283)
- Induced by base substitutions, insertions, deletions and frameshift mutations at the splice acceptor site, donor site or in intronic noncoding regions adjacent to the splice acceptor site (JCO Precis Oncol 2021;5:PO.20.00516, Histopathology 2021;78:1043)
- Kinase domain mutations:
- Mutations in the kinase domain (exons 15 - 21) in MET that lead to upregulation of signaling activity (Oncogene 2004;23:5387)
- Gene fusions:
- Chromosomal rearrangements resulting in a chimeric protein containing at least the kinase domain of MET (Nat Med 2016;22:1314)
- Fusions containing only the MET kinase domain (exon 15 - 21) show much higher downstream activity than fusions containing the full coding region (Nat Med 2016;22:1314)
- Overexpression:
- Highly expressed in a large number of malignancies, including ~50% of lung adenocarcinomas (Clin Cancer Res 2016;22:3048)
- May result from (hypoxia induced) transcriptional activation or MET amplification (Ther Adv Med Oncol 2011;3:S21)
- Poor correlation with amplification and exon 14 skipping (J Thorac Oncol 2019;14:1666)
Clinical features
- In non small cell lung cancer (NSCLC):
- Amplification:
- Low level amplification and polysomy: co-occur with EGFR or KRAS mutations in 2 - 5% of NSCLC (Onco Targets Ther 2020;13:2491, Clin Cancer Res 2016;22:3048)
- High level amplification: ~1% of lung adenocarcinomas, mutually exclusive with other drivers (Clin Cancer Res 2016;22:3048)
- Associated with poor prognosis (Clin Cancer Res 2016;22:3048)
- Poor response to MET inhibitors in primary MET amplified NSCLC (capmatinib, crizotinib) (N Engl J Med 2020;383:944, Ann Oncol 2019;30:1985)
- Most frequent resistance mechanism (7 - 50%) to EGFR inhibitors in EGFR mutant patients (Science 2007;316:1039, Cancer 2020;126:373, Br J Cancer 2019;121:725)
- Resistance mechanism to second or third generation ALK inhibitors in ALK rearranged patients (Clin Cancer Res 2020;26:2535)
- EGFR mutant patients with treatment induced MET amplification benefit from dual EGFR / MET inhibition (Lancet Oncol 2020;21:373, Lancet Respir Med 2020;8:1132)
- ALK rearranged patients with treatment induced MET amplification may benefit from adding / switching to crizotinib (Clin Cancer Res 2020;26:2535)
- Exon 14 skipping:
- Occurs in adenocarcinoma (2.6%), adenosquamous carcinoma (4.8%) and sarcomatoid carcinoma of the lung (31.8%) (Clin Cancer Res 2016;22:3048)
- Associated with poor prognosis (Clin Cancer Res 2016;22:3048)
- Mutually exclusive with other driver mutations, suggesting its role as a primary oncogenic driver (Clin Cancer Res 2016;22:3048)
- Actionable with targeted MET inhibitors, such as capmatinib and tepotinib (N Engl J Med 2020;383:944, N Engl J Med 2020;383:931)
- Gene fusions:
- Extremely rare in lung adenocarcinoma (~0.1%) (cBioPortal for Cancer Genomics: AACR Project GENIE cBioPortal [Accessed 18 August 2021])
- Limited evidence suggests actionability with crizotinib (Clin Cancer Res 2018;24:1337)
- Kinase domain mutations:
- Extremely rare in lung adenocarcinoma (~0.1 - 0.2%) (cBioPortal for Cancer Genomics: AACR Project GENIE cBioPortal [Accessed 18 August 2021])
- Reported as a potential resistance mechanism to crizotinib and osimertinib (Clin Cancer Res 2020;26:2615, Clin Cancer Res 2020;26:2654)
- Limited preclinical evidence suggests actionability with tepotinib and savolitinib (Mol Cancer Ther 2013;12:2415, Clin Cancer Res 2020;26:2615)
- Overexpression:
- Associated with poor prognosis but ineffective as a predictive biomarker for targeted therapy efficacy (Sci Rep 2016;6:35770, J Thorac Oncol 2019;14:1666)
- Amplification:
- In other solid tumors:
- Colorectal adenocarcinoma:
- MET amplifications is detected in 4.4% of patients (Hum Pathol 2018;77:108)
- Gastric carcinoma:
- MET amplification is detected in ~10% of patients (Cancer Epidemiol Biomarkers Prev 2011;20:1021, Br J Cancer 2014;110:1169)
- Glioma:
- MET fusions, especially PTPRZ1-MET, are found in 3 - 7% of pediatric glioblastomas (Cell 2013;155:462)
- Head and neck squamous cell carcinoma:
- Activating MET Y1253D mutation is found in 14% of patients (Nat Genet 1997;16:68)
- Papillary renal cell carcinoma (PRCC):
- MET alterations (activating mutations or amplification) are frequent in type 1 (33%) and type 2 (7%) papillary renal cell carcinomas (Eur Urol 2018;73:71)
- Germline mutations in MET predispose hereditary PRCC (Nat Genet 1997;16:68)
- Other solid tumors:
- MET overexpression is frequent in various solid tumors but is a universally poor predictor of response to MET targeted therapies in the absence of a MET related driver (Nat Rev Clin Oncol 2020;17:569)
- Colorectal adenocarcinoma:
Interpretation
- IHC: membranous and cytoplasmic staining (Onco Targets Ther 2016;9:5809)
- IHC is a poor screening tool for amplification and exon 14 skipping (J Thorac Oncol 2019;14:1666)
Uses by pathologists
- MET IHC correlates poorly with amplifications and exon 14 skipping and therefore has no practical use for pathologists (J Thorac Oncol 2019;14:1666)
- NGS or FISH testing for acquired MET amplification can identify EGFR mutant or ALK rearranged patients who may benefit from second line dual EGFR / MET or ALK / MET inhibition (Lancet Oncol 2020;21:373, Clin Cancer Res 2020;26:2535)
- Multiplex RNA sequencing or NGS can detect exon 14 skipping in NSCLC patients who may benefit from MET inhibitors
Prognostic factors
- Primary high level amplification and exon 14 skipping predict poor prognosis in NSCLC (Clin Cancer Res 2016;22:3048)
Molecular / cytogenetics description
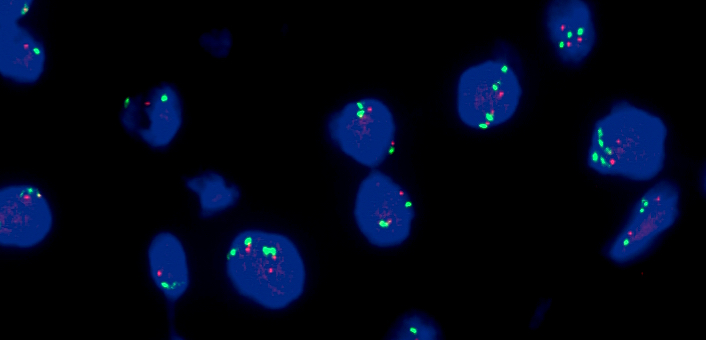
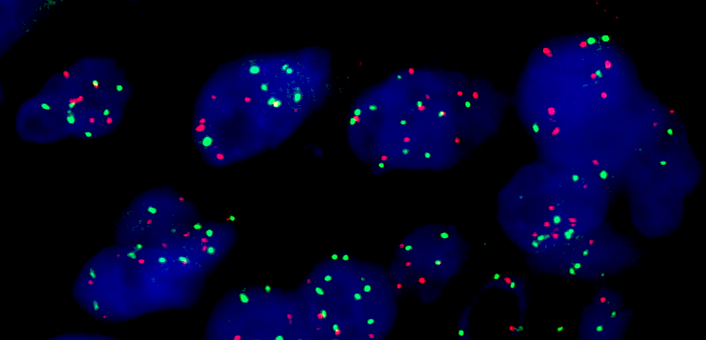
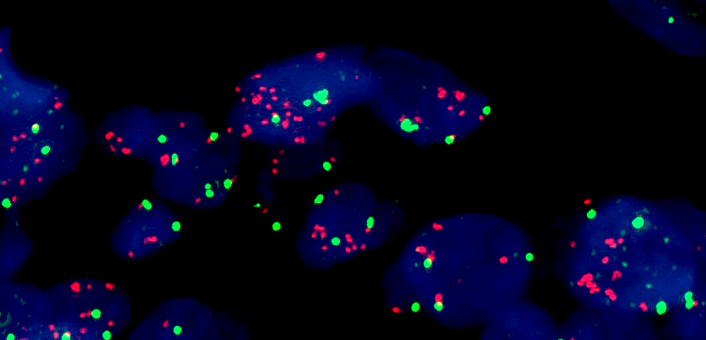
- Fluorescence in situ hybridization
- Gold standard for detection of MET amplification
- Fluorescent probes target MET gene and the centromere of chromosome 7 (CEP7)
- Based on MET gene copy number (per cell) or MET/CEP7 ratio
- Interpretation varies per laboratory; no international consensus on cut off values (J Thorac Oncol 2017;12:15, Nat Rev Clin Oncol 2020;17:569)
- Examples of cut off values for high amplification used by retrospective analyses and clinical trials:
- MET gene copy number ≥ 5 (Cappuzzo criteria) (J Clin Oncol 2009;27:1667)
- MET gene copy number ≥ 6 (Ann Oncol 2020;31:789)
- MET gene copy number ≥ 10 (N Engl J Med 2020;383:944)
- > 15 copies of MET signals in > 10% of tumor cells (J Thorac Oncol 2010;5:305)
- MET/CEP7 ratio ≥ 2 (J Thorac Oncol 2010;5:305)
- MET/CEP7 ratio ≥ 5 (J Thorac Oncol 2016;11:1293)
- Small gene clusters (4 - 10 copies) or innumerable tight gene clusters in > 10% of tumor cells (J Thorac Oncol 2010;5:305)
- Using only gene copy number does not distinguish between true amplification and polysomy (multiple copies of chromosome 7)
- Next generation sequencing (NGS)
- Limited sensitivity / specificity to detect amplification; suspected copy number gain requires FISH confirmation (Nat Rev Clin Oncol 2020;17:569, BMC Cancer 2020;20:291)
- Unbiased detection of MET exon 14 skipping mutations
- Coverage required of the splice acceptor site, donor site and intronic regions adjacent to the splice acceptor (Cancer Discov 2015;5:850)
- Highly variable mechanisms which may include substitutions, insertions, deletions and frameshift mutations (JCO Precis Oncol 2021;5:PO.20.00516, Histopathology 2021;78:1043)
- Detection of a confirmed exon 14 skipping inducing DNA mutation may not require confirmation at RNA level
- Detection of an unknown mutation affecting the exon 14 skipping hotspot regions should prompt an RNA based confirmation test
- RNA analysis
- Enables directly confirming the expression of an exon 13 - exon 15 fused mRNA transcript or MET fusions
- Available techniques include anchored multiplex PCR (Invitae Archer® FusionPlex), targeted RNA sequencing (Asuragen QuantideX®) and molecular counting using fluorescent probes (NanoString nCounter®), among others (J Thorac Oncol 2019;14:737, J Mol Diagn 2019;21:352, Mol Oncol 2021;15:350)
- Multiplex RNA sequencing techniques allow concurrent detection of MET exon 14 skipping, as well as (potentially) actionable fusions involving ALK, BRAF, FGFR1, FGFR2, FGFR3, MET, NRG1, NTRK1, NTRK2, NTRK3, RET and ROS1 in NSCLC
Molecular / cytogenetics images
Sample pathology report
- Stage IV NSCLC with EGFR c.2236_2250del p.(E746_A750del) mutation
- Patient presenting with progressive disease during treatment with an EGFR inhibitor
- Biopsy: metastases in the soft tissue of the os ilium
- Requested: mutation analysis to identify mechanism of resistance and options for targeted therapy
- Result of DNA mutation analysis via next generation sequencing:
- Percentage of tumor cells in the biopsy for DNA extraction: 30%
- Two mutations detected: EGFR (NM_005228.3): c.2236_2250del p.(E746_A750del), 27% variant allele frequency and MET (NM_000245.4): c.2888-44_2899delinsA p.(?), 33% variant allele frequency
- No indication of MET amplification
- Result of RNA analysis to confirm MET exon 14 skipping
- Percentage of tumor cells in the biopsy for RNA extraction: 30%
- MET exon 14 skipping transcripts detected
- Result of DNA FISH analyses to detect MET amplification
- Area for FISH analysis indicated by the pathologist
- Number of MET copies per cell: 1.5
- MET/CEP7 ratio: 0.8
- No amplification of MET
- Conclusion: In addition to the EGFR exon 19 deletion mutation, a MET exon 14 skipping mutation and transcript was detected in this biopsy. In general, patients with this combination of mutations respond to treatment with dual inhibition of EGFR and MET.
- Methodology: Selection of area containing neoplastic cells and estimation of percentage of neoplastic cells in that region was performed by a pathologist. DNA extraction from formalin fixed and paraffin embedded tissue was performed with the Maxwell CSC system (CE-IVD, Promega). Next generation sequencing was performed with the TSO500 panel (Illumina) on a NextSeq platform (Illumina). A > 100 unique coverage of > 98% of all coding regions is required for the detection of a 5% variant allele frequency with more than 95% confidence in tissue with at least 20% neoplastic cells. These quality criteria were met unless stated otherwise. Interpretation of pathogenicity and actionability was according to ACMG and AMP guidelines (J Mol Diagn 2017;19:4, Genet Med 2015;17:405). RNA was extracted with the Maxwell CSC system. RNA sequencing was performed using Archer Fusionplex (SK0133, Invitae) on a MiSeq system (Illumina). Data was analyzed with the Archer analysis tool 6.2. MET FISH analysis on FFPE tissue was performed with the MET (Vysis MET SpectrumRed FISH Probe [06N05-020] and the Vysis CEP 7 [D7Z1] SpectrumGreen Probe [06J37-007]) and counterstained with DAPI. FISH signals were counted in at least 50 neoplastic cells in by 2 independent observers. The laboratory is regulated under NEN-EN-ISO15189 as qualified to perform high complexity testing.
Board review style question #1
A 63 year old man presents with primary adenocarcinoma of the lung with radiological imaging suggestive of metastases to the bones, liver and lymph nodes. Molecular analysis of a lung biopsy (with 60% tumor cells) using next generation sequencing shows no mutations in EGFR, KRAS or BRAF. However, a splice site mutation is detected in MET [NM_000245.4]: c. 2888-30_2898del p.(?) with 42% variant allele frequency. To our knowledge, this specific variant has not been previously described. What is your next step?
- No additional test; report the detected mutation in the pathology report
- Perform an RNA analysis
- Perform fluorescent in situ hybridization (FISH)
- Perform MET immunohistochemistry
- Recommend treatment with a MET inhibitor
Board review style answer #1
B. Perform an RNA analysis. This is a deletion mutation within the splice site region of MET intron 13 - exon 14. However, this is not a previously described MET exon 14 skipping inducing mutation. Therefore, confirmation of the expression of an exon 13 - exon 15 fused mRNA transcript is necessary. This can be done using an RNA based analysis.
Comment Here
Reference: MET
Comment Here
Reference: MET