Table of Contents
Definition / general | Essential features | Terminology | Epidemiology | Sites | Pathophysiology | Laboratory | Treatment | Clinical images | Gross description | Microscopic (histologic) description | Microscopic (histologic) images | Molecular / cytogenetics description | Differential diagnosis | Additional references | Board review style question #1 | Board review style answer #1Cite this page: Lieberman, J.A. Staph coagulase negative. PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/microbiologystaphylococcus.html. Accessed March 30th, 2025.
Definition / general
- Gram stain: positive
- Morphology: cocci in clusters
- Facultative anaerobe
- Part of normal flora of the skin and mucus membranes
Essential features
- Unlike S. aureus, coagulase test is negative
Terminology
- Greek for staphyle ("bunch of grapes") and kokkos ("berry") (Emerg Infect Dis 2013;19:1553)
- Coagulase negative staphylococci (CoNS)
- Many recognized species
Epidemiology
- Important species include Staphylococcus epidermidis, S. saprophyticus, S. lugdunensis and S. haemolyticus
- Common contaminants; to distinguish from true infection rely on clinical judgment, patient features (e.g. hematologic malignancy / immunosuppression) and number / site (central venous catheter vs. venipuncture) of positive cultures (Eur J Clin Microbiol Infect Dis 2012;31:2639, Clin Infect Dis 2004;39:333)
Sites
- Blood stream infections
- Immunocompromised and newborns
- Associated with central venous catheters and other medical devices (Clin Microbiol Rev 2014;27:870)
- Neonatal bloodstream infections / sepsis, lower mortality than S. aureus or Gram negative bacilli (Arch Dis Child Fetal Neonatal Ed 2003;88:F89)
- Urinary tract infections (S. saprophyticus) (Clin Microbiol Rev 2014;27:870)
- Endocarditis (especially S. lugdunensis) (Eur Heart J Acute Cardiovasc Care 2014;3:275)
- Prosthetic joint infections (Int J Artif Organs 2014;37:414)
Pathophysiology
- Biofilm formation and adherence to indwelling devices, catheters
Laboratory
- Culture conditions
- Blood agar plates (nonselective)
- Enrichment broth (tryptic soy broth) in parallel increases culture yield / organism recovery
- Chromogenic agars
- Temperature: 34 - 37 °C, with most species producing growth within 24 hours; a few CoNS, such as S. lentus, may require up to 36 hours for colonies to appear
- Gram positive
- Catalase positive
- Coagulase negative
- Readily identified by matrix associated laser desorption ionization - time of flight mass spectrometry (MALDI-TOF MS)
- MALDI-TOF MS is an increasingly essential tool in the clinical microbiology laboratory for rapid detection of microorganisms
- Small colony variants (SCV) (Clin Microbiol Rev 2016;29:401)
- Especially S. epidermidis, S. capitis and S. warneri
- SCVs more often in foreign body associated infections, chronic or relapsing infections
- Represent metabolic changes, adaptation to intracellular growth
- Ferment fructose
- S. saprophyticus resistant to novobiocin
Treatment
- Beta lactamase resistance and methicillin resistance are widespread
- Vancomycin for methicillin resistant
- Nafcillin for methicillin susceptible
- Presence of mecA and altered PBP2a (penicillin binding protein)
- High rates of resistance to fluoroquinolones, clindamycin, aminoglycoside and macrolides in hospital associated strains; linezolid resistance rare (Antimicrob Agents Chemother 2014;58:1404)
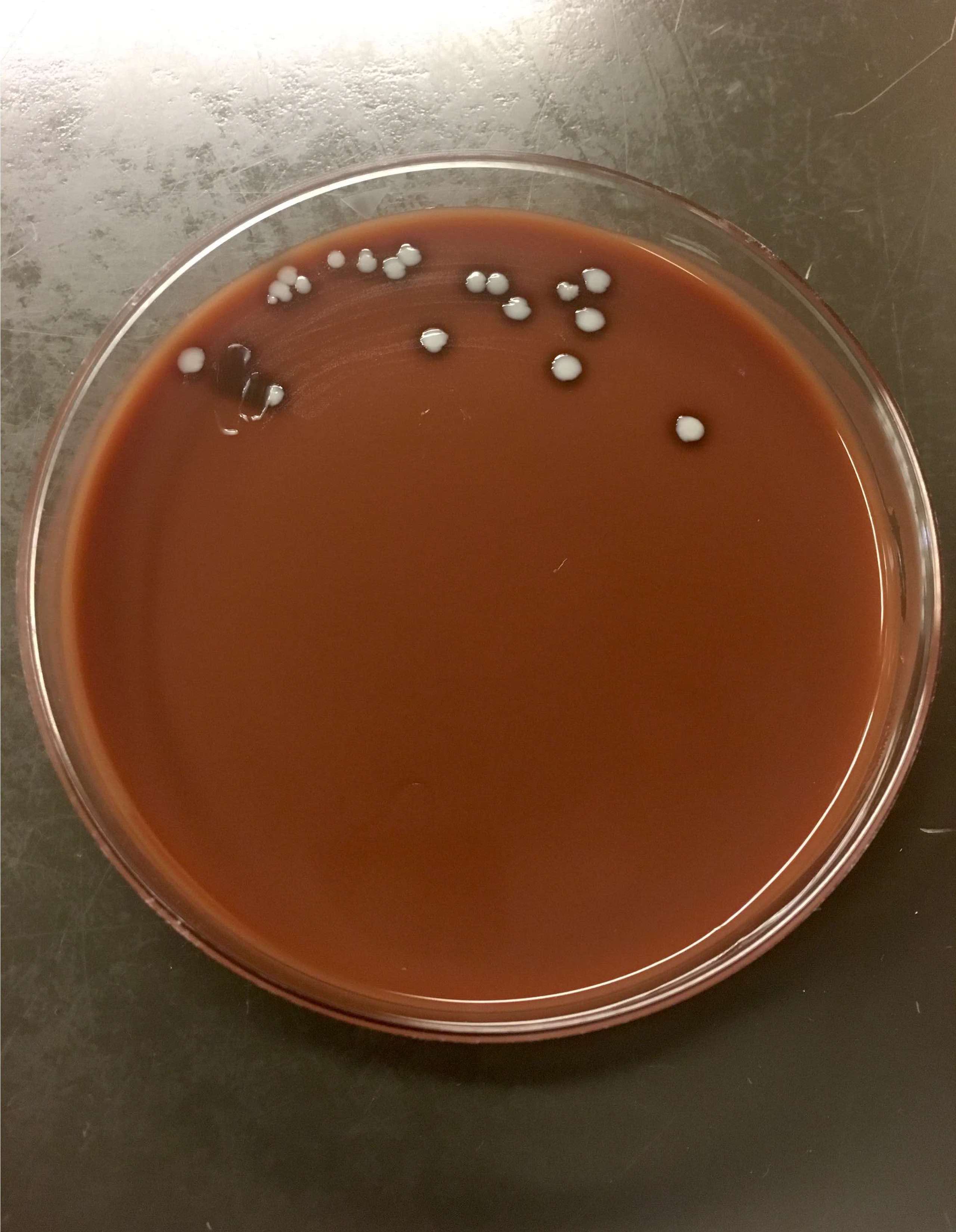
Gross description
- Colonies typically small, beige to white
- Variable hemolysis, depending upon species
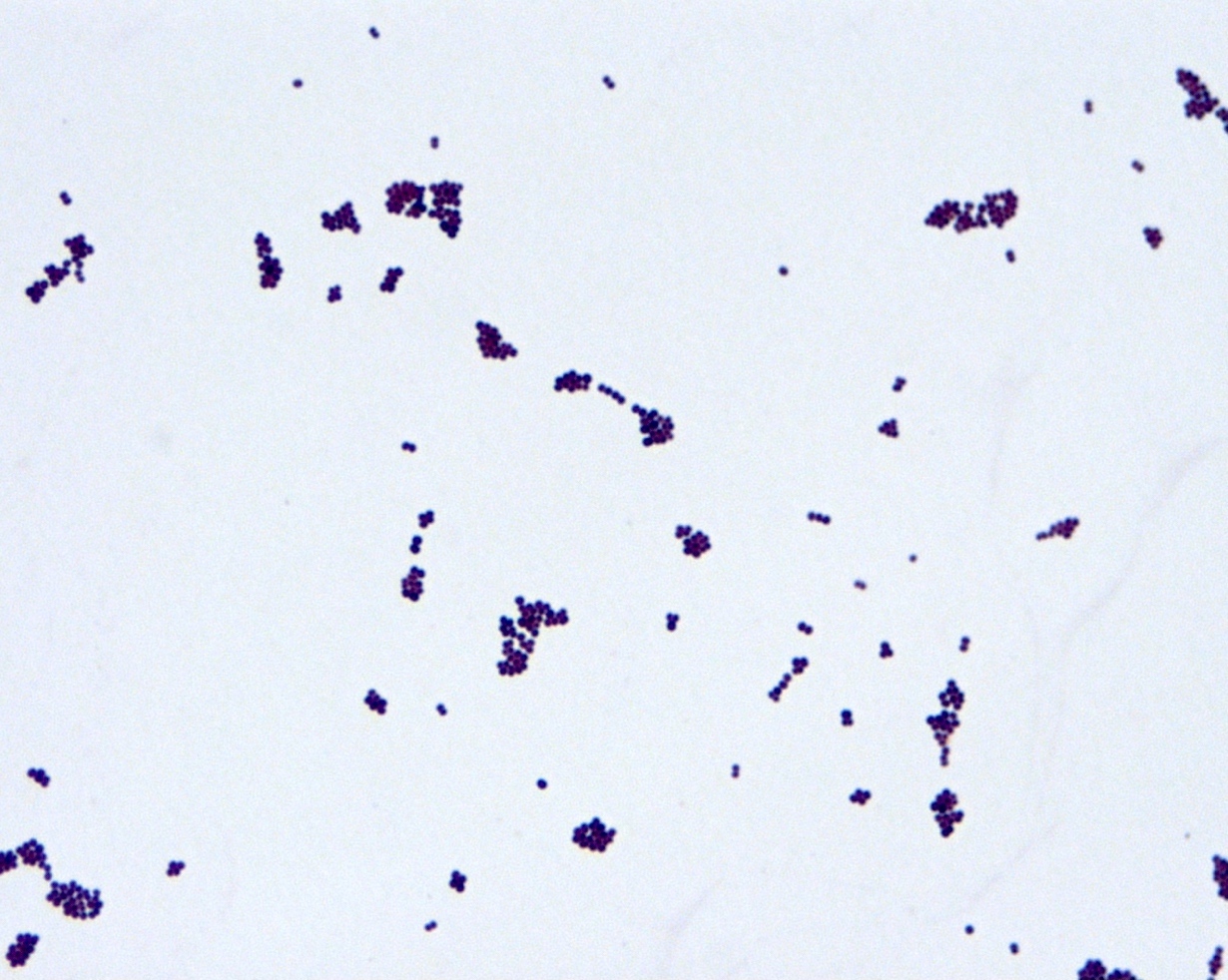
Microscopic (histologic) description
- Gram positive cocci in clusters, similar to other staphylococci
Molecular / cytogenetics description
- Commercial polymerase chain reaction (PCR) based methods, including mecA detection (J Clin Microbiol 2013;51:2072)
- Broad range 16S PCR followed by sequencing and bioinformatic identification
Differential diagnosis
- Primarily other staphylococci
- Aerococcus
- Micrococcus
Additional references
Board review style question #1
Board review style answer #1
A. Altered penicillin binding protein. The image shows Gram positive cocci in clusters. PBP2a, an altered (low affinity) penicillin binding protein, is the product of the mecA gene, and mediates oxacillin / methicillin resistance in staphylococci. Answer B is incorrect because although many Staphyloccoci strains are resistant to a variety of beta-lactam antibiotics, the extended spectrum beta lactamases (ESBLs) are a specific class of enzymes that destroy a wide range of beta-lactam antibiotics, and these are found in Gram-negative pathogens, particularly Enterobacteriaceae. Answer C is incorrect because mutations in DNA gyrase confer resistance to quinolone / fluoroquinolone antibiotics. Answer D is incorrect because point mutations in the 23S rRNA gene confer macrolide resistance, especially in Helicobacter pylori, by altering drug binding sites. Answer E is incorrect because porins are outer membrane proteins only found in Gram negative organisms. Porin mutations can block or reduce drug influx and drug effect; examples include Acinetobacter baumanii and imipenem and Pseudomonas aeruginosa and tobramycin.
Comment Here
Reference: Staph coagulase negative
Comment Here
Reference: Staph coagulase negative