Table of Contents
Definition / general | Essential features | Terminology | Applications | Implementation | Advantages | Limitations | Software | Diagrams / tables | Board review style question #1 | Board review style answer #1 | Board review style question #2 | Board review style answer #2Cite this page: Levy J, Le MK, Vaickus L. Application of graph neural networks to whole slide images . PathologyOutlines.com website. https://www.pathologyoutlines.com/topic/informaticsgraphneuralnetworkstowholeslideimages.html. Accessed March 31st, 2025.
Definition / general
- Graphs in biological context: graphs represent biological entities like tissue regions or cells as nodes with specific attributes or features such as histological, molecular or textual data; edges between these nodes signify relationships based on aspects like similar expression patterns or spatial proximity
- Function of graph neural networks (GNN): GNNs facilitate information exchange between nodes, providing context to objects of interest based on their neighboring attributes; this aids in understanding intricate biological phenomena, such as tissue structure interactions or cellular communication
- Uniqueness and applicability of GNNs: GNNs capture tissue and spatial molecular features across different scales for predictive tasks; while other graph based algorithms and statistical methods exist, GNNs stand out by harnessing the power of deep learning, enabling intricate analyses over extensive patient data sets
Essential features
- Whole slide images (WSIs) are digital versions of tissue samples that can vary widely in size, scale and orientation; these variations pose challenges for computer vision algorithms to effectively analyze histological features across different scales
- Graphs are flexible data structures that can represent complex interactions, such as those between cells in whole slide images; in these graphs, cells and patch based grids are represented as nodes, histological characteristics as node attributes and spatial relationships between adjacent patches / cells as edges
- Nodal attributes are often derived using convolutional neural networks focused on different tasks; when using cell graph neural networks (CGNNs), cell detection algorithms are necessary to gather cell features
- GNNs excel at processing spatial information (e.g., in identifying tissue types, GNNs can evaluate neighboring tissue regions to make context sensitive decisions)
- GNNs are used for various tasks including node classification / regression (e.g., tissue segmentation, gene / protein expression prediction), graph classification (integrating whole slide information), clustering / self supervised learning (identifying tissue domains) and multimodal approaches (combining data types for improved prognostication)
Terminology
- Whole slide image (WSI): digitized representation of histologic slide after whole slide scanning at 20x or 40x resolution (e.g., using Aperio AT2 or GT450 scanner); slide dimensionality can exceed 100,000 pixels in any given spatial dimension and typically contains 3 color channels: red, green and blue (RGB) (Annu Rev Pathol 2013;8:331)
- Subimage / patch: smaller, local rectangular region extracted from a whole slide image, often done to reduce the computational resources required for the development and deployment of machine learning algorithms (J Pathol Inform 2019;10:9)
- Graphs: mathematical constructs that represent biological entities as nodes, their features as attributes and relationships between the nodes as edges (Pac Symp Biocomput 2021;26:285)
- Whole slide graph (WSG): representation of whole slide images using a graph data structure based on spatial proximity between detected cells or evenly spaced subimages; nodal attributes can be represented through various methods (e.g., positive / negative staining of various markers from multiplexed immunofluorescence for specific cells)
- Artificial intelligence: computational approaches developed to perform tasks that typically require human intelligence / semantic understanding (Lancet Oncol 2019;20:e253)
- Machine learning: computational heuristics that learn patterns from data without requiring explicit programming to make decisions (see Artificial intelligence) (Med Image Anal 2016;33:170)
- Classification, regression, self supervision, clustering, dimensionality reduction and detection
- Evaluation metrics (accuracy, AUC, F1, sensitivity, specificity and intersection over union [IoU])
- Training, validation and test cohort
- Deep learning: a group of machine learning algorithms that specifically consist of artificial neural networks that contain multiple hidden layers, typically ranging from dozens to hundreds, depending on the complexity of the problem at hand (Nature 2015;521:436)
- Artificial neural networks (ANN): a type of machine learning algorithm that represents input data (e.g., images) as nodes (neurons); learns image filters (e.g., color, shapes) used to extract histomorphological / cytological features; comprised of multiple processing layers to represent objects at multiple levels of abstraction (deep learning); inspired by the visual cortex (Nature 2015;521:436)
- Convolutional neural networks: ideal for image data (e.g., whole slide images), work by storing filters to extract task relevant shapes and patterns
- Graph neural networks: type of deep learning model that shares information between neighboring nodes using message passing operations; features convolutional, pooling and attention layers
- Convolutional layers primarily share information between neighbors to provide additional context
- Pooling layers simplify the graph structure by summarizing various cellular / tissue compartments (e.g., a single node to represent many nodes from the submucosal layer of the colon)
- Attention layers dynamically decide how much information to incorporate from the node's neighbors or across the nodes of a graph based on their relevance for a specific prediction; can be integrated into convolutional or pooling layers
- Multiple instance learning (MIL): a type of supervised learning where a group of instances (such as images, cells, subregions) is labeled without labeling individual instances; in MIL, a group of related instances with one label is called a bag and in the context of computational pathology, a bag usually corresponds to a whole slide image, while constituent image patches represent instances inside the bag
- Weakly supervised learning: not every individual instance is actually labeled
- Graph data structures (Nat Biomed Eng 2022;6:1353)
- Nodes: entities or objects to be related
- Edges: describe relationship between pairs of nodes
- Nodal attributes: numerical characteristics that describe each node
- Edge attributes: numerical characteristics that describe each relationship
- Adjacency matrix: matrix providing information about the presence of a relationship between all pairs of nodes
Applications
- Examples of GNNs applied to whole slide images
- Early applications (Nat Commun 2022;13:4250, MICCAI 2018;11071:174, Med Image Anal 2022;75:102264, ICCV 2019;0)
- SlideGraph+ (Med Image Anal 2022;80:102486, IEEE 2020;1049)
- Additional reviews (Comput Med Imaging Graph 2022;95:102027, Nat Biomed Eng 2022;6:1353)
- Node level prediction tasks
- Delineating tissue subcompartments for colon cancer staging (Pac Symp Biocomput 2021;26:285)
- Intraoperative margin assessment (Exp Dermatol 2024;33:e14949, JAAD Int 2024;15:185, NPJ Precis Oncol 2024;8:2)
- Placenta villous structure (Nat Commun 2024;15:2710)
- Immune cell prediction (PMLR 2022;194:15)
- Basal cell carcinoma (GLMI 2019;11849:112)
- Multiplex immunofluorescence (PNAS Nexus 2023;2:pgad171, Nat Rev Cancer 2023;23:508)
- Graph level prediction
- Recurrence prediction (NPJ Digit Med 2023;6:48)
- Lymph node metastasis (CVPR 2020;4837)
- Histologic subtyping (Arch Pathol Lab Med 2023;147:1251, J Pathol Inform 2022;13:100158)
- Tumor mutation burden (Am J Pathol 2023;193:2111)
- Tumor stage (Pac Symp Biocomput 2021;26:285)
- Glioblastoma (GBM) prognostication (Nat Commun 2023;14:4122)
- Multiplexed immunofluorescence (mIF) (Nat Commun 2023;14:4122)
- Fibrosis staging (CVPR 2022;1835)
- Chronic kidney disease (Sci Rep 2023;13:12701)
- Multimodal approaches
- Prediction of spatial transcriptomics (ST) from whole slide images (Brief Bioinform 2022;23:bbac297, J Pathol Inform 2023;14:100308)
- Clustering spatial transcriptomics and whole slide images (Cell Syst 2023;14:404, Nat Commun 2023;14:1155, Nat Methods 2021;18:1342)
- Survival outcomes (Pac Symp Biocomput 2024;29:464, BioData Min 2023;16:23, IEEE Trans Med Imaging 2022;41:757, SAC '22: Proceedings of the 37th ACM/SIGAPP Symposium on Applied Computing 2022;636)
- Interpretation
- Identification of prognostic graph features (Nat Biomed Eng 2022 Aug 18 [Epub ahead of print])
- Inflammatory bowel disease (IBD) (Inflammatory Bowel Diseases 2023;29:S22)
- Region of interest retrieval (MICCAI 2019;11764:550, IEEE 2020;1049)
- Histocartography (CVPR 2021;8106)
- Subgraphs (Lancet Digit Health 2022;4:e787, Cancer Res 2022;82:1922)
- Early applications (Nat Commun 2022;13:4250, MICCAI 2018;11071:174, Med Image Anal 2022;75:102264, ICCV 2019;0)
Implementation
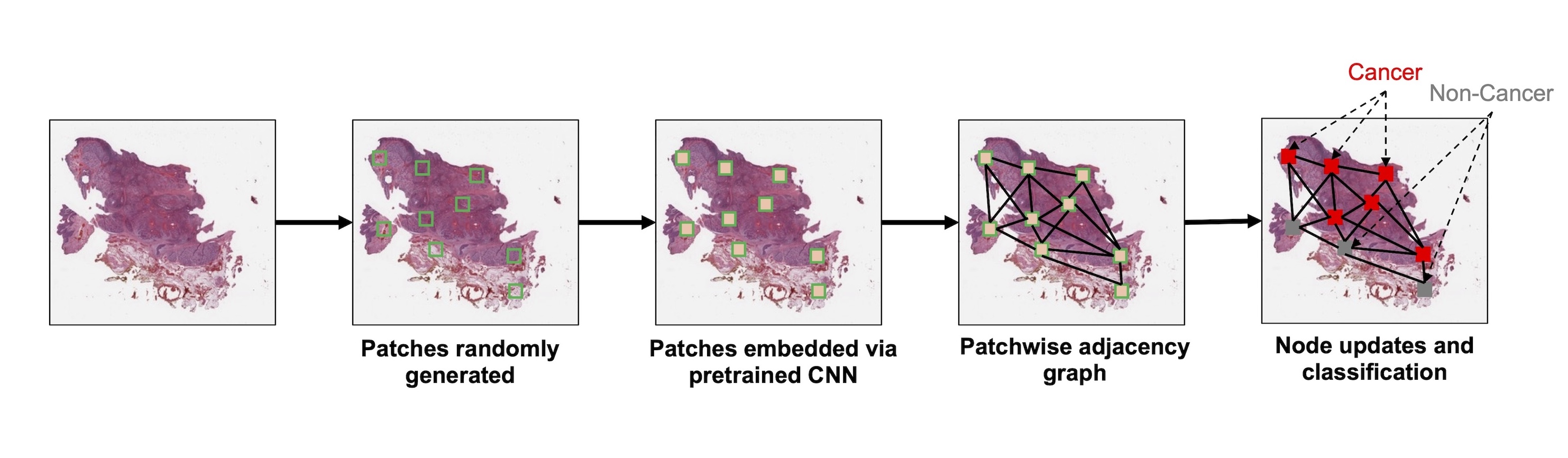
- Preparing graph structured data from whole slide images
- Tissue preprocessing: using various image processing techniques to isolate large contiguous regions of tissue
- Nodes: patches, nuclei / cells, clustered microarchitectures / tissue regions
- Graph formation based on breaking down tissue into various constituent components (i.e., nodes)
- Localized cells: use of cell detection / segmentation algorithms (e.g., UNET, YOLOv8, Detectron, Mask-RCNN, hematoxylin deconvolution) to assign positional coordinates to cells throughout a slide
- Underappreciates diagnostic / prognostic information contained outside of cell dense regions (e.g., large scale interactions with fibrous stroma)
- Highly dependent on cell detection accuracy and information extracted from cells, very complex and computationally intractable based on number of cells
- Farthest point sampling reduces complexity
- Tissue microarrays can also make cell graphs possible but can introduce sampling bias
- Tissue patches
- Patches can also be captured based on partitioning of tissue through segmentation based / superpixel approaches (e.g., SLIC) or regularly spaced
- Localized cells: use of cell detection / segmentation algorithms (e.g., UNET, YOLOv8, Detectron, Mask-RCNN, hematoxylin deconvolution) to assign positional coordinates to cells throughout a slide
- Graph formation based on breaking down tissue into various constituent components (i.e., nodes)
- Edges: typically based on spatial proximity (e.g., k nearest neighboring cells or patches within a 50 micron radius)
- Nodal attributes / embeddings: usually a set of features derived from applying a convolutional neural network (CNN) or vision transformer to an image associated with the node; can also represent various shape, color and texture features (e.g., gray level co-occurrence matrix, custom image filters, etc.)
- Embeddings depend on the learning objective for CNN, e.g., supervised / unsupervised / self supervised
- Methods to extract cell / patch features to form nodal attributes
- Based on raw imaging features
- Morphological / shape characteristics: texture based analysis (e.g., gray level co-occurrence), nuclei features (e.g., presence of grooves, frayed chromatin), nuclei shape (e.g., eccentricity), etc.
- Multiplexed immunofluorescence / immunohistochemistry (IHC): staining intensities for detected cells
- Convolutional / transformer approaches
- Any combination of the above features
- Based on raw imaging features
- Targets
- Node level: tissue region classification, spatial gene expression patterns, cell type prediction (e.g., immune cell), etc.
- Graph level: prediction of nodal metastasis, various bulk molecular alterations, survival outcomes, etc.
- Input data preprocessing
- Routine tissue staining: utilizes hematoxylin for highlighting nuclei and eosin for visualizing cytoplasm and extracellular matrix
- Immunostaining: detect and visualize specific proteins of interest
- Immunohistochemistry: typically uses 2 to 3 stains through enzyme labeled antibodies, producing chromogenic reactions that deposit colors that can be visualized through brightfield imaging
- Multiplexed immunofluorescence: relies on spectral unmixing to differentiate antibodies conjugated to fluorophores that may spectrally overlap, which are employed for labeling biomolecules like proteins
- Structure of GNNs
- Graph convolution: shares information between neighboring nodes as added context; nodes pass messages along edges and update their states based on received messages
- Graph attention: weighs the importance of nodes differently during convolution
- GraphSAGE: aggregates features from a node's local neighborhood using sampling techniques
- Graph convolutional network (GCN): uses a spectral based approach to define convolutions on graphs
- Pooling methods: techniques to downsample nodes and edges for hierarchical representation; represents objects at higher abstraction; akin to decreasing the spatial resolution of an image to represent more complex phenomena
- DiffPool: differentiable pooling that assigns nodes to clusters in a hierarchical manner
- TopK pooling: retains the top k nodes based on a scoring function
- SAGPool: self attention graph pooling that scores nodes using a self attention mechanism
- Self attention: technique that provides a score for each node based on their relevance
- Data augmentation: techniques to enhance the robustness and generalization of models
- Drop node: randomly removes nodes during training
- Drop edge: randomly removes edges during training
- Subgraph sampling: samples subgraphs to create varied training instances
- Feature masking: randomly masks node attributes during training
- Graph convolution: shares information between neighboring nodes as added context; nodes pass messages along edges and update their states based on received messages
- Examples of types of GNNs for various prediction tasks
- Node level classification / regression: predicting labels or continuous values for individual node
- Examples: automatic annotation, spatial gene expression, cancer cell detection
- Node level clustering: grouping nodes into clusters based on their features and connectivity
- Examples: identifying functional modules in biological networks, customer segmentation in social networks
- Graph level classification / regression: predicting labels or continuous values for entire graphs
- Examples: risk stratification, cancer detection, bulk gene expression
- Graph level clustering: grouping entire graphs into clusters based on their overall structure
- Examples: identification of novel histological variants
- Self supervised learning: learning representations without explicit labels by leveraging the structure of the graph; can help cluster nodes and graphs and reduce the amount of data needed for supervised analysis (Pac Symp Biocomput 2024;29:464, BioData Min 2023;16:23)
- Examples: contrastive learning, node embeddings, graph embeddings
- PyGCL: self supervised contrastive learning (ArXiv: An Empirical Study of Graph Contrastive Learning [Accessed 20 August 2024])
- Graph clustering (ArXiv: Spectral Clustering with Graph Neural Networks for Graph Pooling [Accessed 20 August 2024])
- Examples: contrastive learning, node embeddings, graph embeddings
- Link prediction: predicting the existence or likelihood of edges between nodes
- Examples: recommending friends in social networks, predicting protein - protein interactions
- Graph autoencoder (IJCAI (US) 2018;2609)
- Examples: recommending friends in social networks, predicting protein - protein interactions
- GNN interpretation: explaining and interpreting the predictions made by GNNs; can highlight important nodes, edges, subgraphs, features
- Examples: GNNExplainer, Captum, integrated gradients, attention mechanisms
- Transductive versus inductive: approaches for making predictions
- Transductive: making predictions on specific nodes seen during training
- Inductive: generalizing to unseen instances and graphs beyond the training data
- Node level classification / regression: predicting labels or continuous values for individual node
Advantages
- Motivation for GNNs
- Limitations of traditional machine learning (ML) / convolutional neural networks: traditional machine learning analysis of tissue subdivides tissue into patches and analyzes patches independently, while information is actually shared in adjacent patches and better captured using a graph
- Traditional machine learning does not consider surrounding tissue context
- GNNs relax assumptions made by convolutional neural networks and are more flexible to the tissue shape, orientation, ordering of patches
- Graph based methods consider surrounding tissue context
- Examples of alternative algorithmic / statistical methods for studying graph data (Cancer Res 2020;80:1199, Diagnostic Pathology 2015;1:61)
- Community detection / clustering: partitions a network into groups of highly connected nodes with similar attributes
- Module based methods: Louvain, Leiden
- Statistical approaches: latent ERGM
- Graph embedding: spectral clustering
- Hotspot analysis: Getis-Ord G statistics
- Influence analysis
- Centrality: eigenvector, betweenness, degree
- Network autocorrelation models: captures dependence of how values for an individual point are influenced by characteristics of neighbors
- Community detection / clustering: partitions a network into groups of highly connected nodes with similar attributes
- Prior forms of representing graph structured data in pathology slides
- Graph formation
- Voronoi diagrams: segment regions based on the nearest cell
- Delaunay triangulation: connect cells with triangles, ensuring no cell is inside any triangle's circumcircle
- Subgraph: analyze specific regions or features within the larger graph
- Cell graph: nodes represent cells, edges represent spatial relationships
- Skeletonization: reduce to essential lines and structures (Cancer Res 2024;84:3516)
- Minimum spanning trees: connect all cells with minimum total edge weight
- Spatial neighborhood analysis: calculate the proportions of cell types within a neighborhood to use as features
- Graph features
- Morphological features: cell shapes and structures
- Texture / intensity features: assess cell texture and staining intensity
- Statistical / spectral descriptors: use statistical measures and spectral analysis for cell attributes
- Clustering coefficients: measure the tendency of cells to cluster in local regions
- Size: evaluate cell or cluster sizes
- Degree: count each cell's connections
- Centralization: measures extent to which network is dominated by a few central nodes
- Topological analysis: mapper and persistence homology (Leukemia 2023;37:348, Insights Imaging 2023;14:58, Dev Dyn 2020;249:816, MLMI 2014;8679:231)
- Graph formation
- Alternative methods for studying whole slide image data (Pac Symp Biocomput 2021;26:285)
- Voting based method: counting the number of patches across a slide with various classifications and developing a decision rule through machine learning for the final classification
- Tissue characteristics introduce potentially flawed decision rules that could change outcomes heavily based on tissue sampling
- Attention based methods: patch information is aggregated using a weighted average, inference is somewhat stochastic and does not make any assumptions about the underlying tissue structure or downplay higher order context between patch and neighborhood
- Multiple instance learning (MIL): treating the whole slide image as a bag of instances (patches); the MIL algorithm to aggregate the instances (patches) largely depends on the problem at hand and does not explicitly rely on tissue context
- Treating entire slide as an image
- Places reliance on tissue orientation / positioning, significant computational complexity
- Voting based method: counting the number of patches across a slide with various classifications and developing a decision rule through machine learning for the final classification
- Limitations of traditional machine learning (ML) / convolutional neural networks: traditional machine learning analysis of tissue subdivides tissue into patches and analyzes patches independently, while information is actually shared in adjacent patches and better captured using a graph
Limitations
- Can potentially smooth over information locally and have challenges in capturing sharp discontinuities in the data
- May rely on accurate nuclei detection
- Can require significant computation and difficult to optimize if opting to simultaneously improve both the extraction of numerical characteristics of cells / patches and graph modeling end to end
- For modeling histopathology, tissue may need to remain intact for best performance; sectioning quality may impact performance
- Somewhat limited capability in modeling long range dependencies across WSI unless combined with attention based approaches
- May be suitable for modeling cell clusters in cytology specimens but not individual cells and their connectivity
Software
- Select examples of software for GNN algorithm development
- Scikit-image, OpenCV2: image analysis frameworks (PeerJ 2014;2:e453, Comput Biol Med 2017;84:189)
- Scikit-learn, caret: machine learning framework (Bioinformatics 2023;39:btac829, J Open Source Softw 2019;4:1903)
- PyTorch, Keras, TensorFlow: deep learning frameworks (Mol Cancer Res 2022;20:202)
- Instructional book for developing deep learning workflows: D2L: Dive into Deep Learning, 1st Edition, 2023
- Self supervised learning for convolutional, graph neural networks
- Detectron2, MMDetection: cell detection frameworks (Cancer Cytopathol 2023;131:19)
- Captum, SHAP, GNNExplainer, histocartography: model interpretation frameworks (Nat Mach Intell 2020;2:56, Adv Neural Inf Process Syst 2019;32:9240, ArXiv: Captum - A Unified and Generic Model Interpretability Library for PyTorch [Accessed 20 August 2024], PMLR 2021;156:117)
- GNN libraries: PyTorch Geometric, Spektral, DGL, DeepSNAP (ArXiv: Fast Graph Representation Learning with PyTorch Geometric [Accessed 20 August 2024], IEEE Comput Intell Mag 2021;16:99, ArXiv: Deep Graph Library - A Graph-Centric, Highly-Performant Package for Graph Neural Networks [Accessed 20 August 2024], ACM Trans Intell Syst Technol 2016;8:1)
- Topological data analysis (TDA): giotto-tda, scikit-TDA, Ripser, Kepler Mapper, Deep Graph Mapper (JMLR 2021;22:1, Zenodo: scikit-tda/scikit-tda, v1.1.0 [Accessed 2 August 2024], J Open Source Softw 2018;3:925, J Open Source Softw 2019;4:1315, Front Big Data 2021;4:680535)
Diagrams / tables
Board review style question #1
In a pathology graph constructed from a whole slide image, what is typically represented by nodal attributes?
- Diagnosis associated with each slide
- Metadata of the slide including the date of preparation and the patient's details
- Numerical characteristics describing features such as cell morphology or staining intensity
- Type of scanner used to digitize the whole slide image
Board review style answer #1
C. Numerical characteristics describing features such as cell morphology or staining intensity. Information must be stored at and shared between specific locations in the tissue slide that encompasses the underlying histomorphology / molecular information. Predictive modeling with graph neural networks requires the extraction and sharing of information organized spatially within a slide and could not be accomplished without this information. Answer A is incorrect because the diagnosis is an example of an outcome for a graph level classification task and can be derived from summarizing nodal attributes across the patches in a slide. Answer B is incorrect because metadata are patient level characteristics and do not represent local, patch / cellwise imaging features. Answer D is incorrect because the type of scanner is unrelated to the nodal attributes in the context of whole slide graphs.
Comment Here
Reference: Application of graph neural networks to whole slide images
Comment Here
Reference: Application of graph neural networks to whole slide images
Board review style question #2
What is the primary function of convolutional layers in a graph neural network (GNN) when applied to pathology images?
- To adjust the color balance of histology images for better visualization
- To classify the entire slide image into benign or malignant
- To encrypt patient data for secure transmission
- To share information between neighboring nodes, providing context to biological entities
Board review style answer #2
D. To share information between neighboring nodes, providing context to biological entities. Graph convolution operators are used to capture spatial dependencies between adjacent tissue regions / cells to improve predictive modeling above and beyond modeling this information independently without graph convolutions. Answer A is incorrect because adjusting the color balance or normalizing the tissue stain can help improve the predictive accuracy of the algorithm; however, this task is typically performed during preprocessing and not by convolutional layers. Answer B is incorrect because classifying entire slide images is generally achieved through pooling layers that aggregate information at a higher level. Answer C is incorrect because data encryption is unrelated to the function of convolutional layers in GNNs.
Comment Here
Reference: Application of graph neural networks to whole slide images
Comment Here
Reference: Application of graph neural networks to whole slide images